Diabetes Metab J.
2018 Aug;42(4):270-281. 10.4093/dmj.2018.0101.
Role of the Pyruvate Dehydrogenase Complex in Metabolic Remodeling: Differential Pyruvate Dehydrogenase Complex Functions in Metabolism
- Affiliations
-
- 1Leading-edge Research Center for Drug Discovery and Development for Diabetes and Metabolic Disease, Kyungpook National University Hospital, Daegu, Korea. leei@knu.ac.kr, smpark93@gmail.com
- 2Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Korea.
- 3Department of Biomedical Science & BK21 plus KNU Biomedical Convergence Programs, Kyungpook National University, Daegu, Korea.
- KMID: 2418715
- DOI: http://doi.org/10.4093/dmj.2018.0101
Abstract
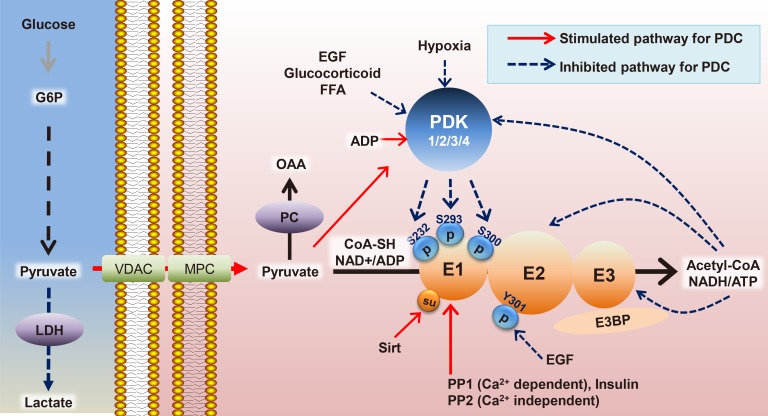
- Mitochondrial dysfunction is a hallmark of metabolic diseases such as obesity, type 2 diabetes mellitus, neurodegenerative diseases, and cancers. Dysfunction occurs in part because of altered regulation of the mitochondrial pyruvate dehydrogenase complex (PDC), which acts as a central metabolic node that mediates pyruvate oxidation after glycolysis and fuels the Krebs cycle to meet energy demands. Fine-tuning of PDC activity has been mainly attributed to post-translational modifications of its subunits, including the extensively studied phosphorylation and de-phosphorylation of the E1α subunit of pyruvate dehydrogenase (PDH), modulated by kinases (pyruvate dehydrogenase kinase [PDK] 1-4) and phosphatases (pyruvate dehydrogenase phosphatase [PDP] 1-2), respectively. In addition to phosphorylation, other covalent modifications, including acetylation and succinylation, and changes in metabolite levels via metabolic pathways linked to utilization of glucose, fatty acids, and amino acids, have been identified. In this review, we will summarize the roles of PDC in diverse tissues and how regulation of its activity is affected in various metabolic disorders.
MeSH Terms
-
Acetylation
Amino Acids
Citric Acid Cycle
Diabetes Mellitus, Type 2
Fatty Acids
Glucose
Glycolysis
Metabolic Diseases
Metabolic Networks and Pathways
Metabolism*
Mitochondria
Neurodegenerative Diseases
Obesity
Oxidative Phosphorylation
Oxidoreductases
Phosphoric Monoester Hydrolases
Phosphorylation
Phosphotransferases
Protein Processing, Post-Translational
Pyruvate Dehydrogenase Complex*
Pyruvic Acid*
Amino Acids
Fatty Acids
Glucose
Oxidoreductases
Phosphoric Monoester Hydrolases
Phosphotransferases
Pyruvate Dehydrogenase Complex
Pyruvic Acid
Figure
Cited by 1 articles
-
Progression of Multifaceted Immune Cells in Atherosclerotic Development
Sungmi Park, In-Kyu Lee
J Lipid Atheroscler. 2019;8(1):15-25. doi: 10.12997/jla.2019.8.1.15.
Reference
-
1. Stacpoole PW. The pyruvate dehydrogenase complex as a therapeutic target for age-related diseases. Aging Cell. 2012; 11:371–377. PMID: 22321732.
Article2. Jeoung NH, Harris CR, Harris RA. Regulation of pyruvate metabolism in metabolic-related diseases. Rev Endocr Metab Disord. 2014; 15:99–110. PMID: 24214243.
Article3. Muoio DM, Noland RC, Kovalik JP, Seiler SE, Davies MN, DeBalsi KL, Ilkayeva OR, Stevens RD, Kheterpal I, Zhang J, Covington JD, Bajpeyi S, Ravussin E, Kraus W, Koves TR, Mynatt RL. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012; 15:764–777. PMID: 22560225.
Article4. Kankotia S, Stacpoole PW. Dichloroacetate and cancer: new home for an orphan drug? Biochim Biophys Acta. 2014; 1846:617–629. PMID: 25157892.
Article5. Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014; 158:84–97. PMID: 24995980.
Article6. Bowker-Kinley MM, Davis WI, Wu P, Harris RA, Popov KM. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J. 1998; 329(Pt 1):191–196. PMID: 9405293.
Article7. Saunier E, Benelli C, Bortoli S. The pyruvate dehydrogenase complex in cancer: an old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int J Cancer. 2016; 138:809–817. PMID: 25868605.
Article8. Battello N, Zimmer AD, Goebel C, Dong X, Behrmann I, Haan C, Hiller K, Wegner A. The role of HIF-1 in oncostatin M-dependent metabolic reprogramming of hepatic cells. Cancer Metab. 2016; 4:3. PMID: 26889381.
Article9. Huang B, Wu P, Popov KM, Harris RA. Starvation and diabetes reduce the amount of pyruvate dehydrogenase phosphatase in rat heart and kidney. Diabetes. 2003; 52:1371–1376. PMID: 12765946.
Article10. Tso SC, Lou M, Wu CY, Gui WJ, Chuang JL, Morlock LK, Williams NS, Wynn RM, Qi X, Chuang DT. Development of dihydroxyphenyl sulfonylisoindoline derivatives as liver-targeting pyruvate dehydrogenase kinase inhibitors. J Med Chem. 2017; 60:1142–1150. PMID: 28085286.
Article11. Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013; 50:919–930. PMID: 23806337.
Article12. Fukushima A, Alrob OA, Zhang L, Wagg CS, Altamimi T, Rawat S, Rebeyka IM, Kantor PF, Lopaschuk GD. Acetylation and succinylation contribute to maturational alterations in energy metabolism in the newborn heart. Am J Physiol Heart Circ Physiol. 2016; 311:H347–H363. PMID: 27261364.
Article13. Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. 2014; 289:16615–16623. PMID: 24798336.
Article14. Xiangyun Y, Xiaomin N, Linping G, Yunhua X, Ziming L, Yongfeng Y, Zhiwei C, Shun L. Desuccinylation of pyruvate kinase M2 by SIRT5 contributes to antioxidant response and tumor growth. Oncotarget. 2017; 8:6984–6993. PMID: 28036303.
Article15. Zhu D, Hou L, Hu B, Zhao H, Sun J, Wang J, Meng X. Crosstalk among proteome, acetylome and succinylome in colon cancer HCT116 cell treated with sodium dichloroacetate. Sci Rep. 2016; 6:37478. PMID: 27874079.
Article16. Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab. 2015; 3:10. PMID: 26401273.
Article17. Foster DW. Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. J Clin Invest. 2012; 122:1958–1959. PMID: 22833869.
Article18. Dong K, Ni H, Wu M, Tang Z, Halim M, Shi D. ROS-mediated glucose metabolic reprogram induces insulin resistance in type 2 diabetes. Biochem Biophys Res Commun. 2016; 476:204–211. PMID: 27207834.
Article19. Caton PW, Holness MJ, Bishop-Bailey D, Sugden MC. PPARα-LXR as a novel metabolostatic signalling axis in skeletal muscle that acts to optimize substrate selection in response to nutrient status. Biochem J. 2011; 437:521–530. PMID: 21609322.
Article20. Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, Feron O. Acidosis drives the reprogramming of fatty acid metabolism in cancer cells through changes in mitochondrial and histone acetylation. Cell Metab. 2016; 24:311–323. PMID: 27508876.
Article22. Griffin TM, Humphries KM, Kinter M, Lim HY, Szweda LI. Nutrient sensing and utilization: Getting to the heart of metabolic flexibility. Biochimie. 2016; 124:74–83. PMID: 26476002.
Article23. Vadvalkar SS, Baily CN, Matsuzaki S, West M, Tesiram YA, Humphries KM. Metabolic inflexibility and protein lysine acetylation in heart mitochondria of a chronic model of type 1 diabetes. Biochem J. 2013; 449:253–261. PMID: 23030792.24. Mori J, Basu R, McLean BA, Das SK, Zhang L, Patel VB, Wagg CS, Kassiri Z, Lopaschuk GD, Oudit GY. Agonist-induced hypertrophy and diastolic dysfunction are associated with selective reduction in glucose oxidation: a metabolic contribution to heart failure with normal ejection fraction. Circ Heart Fail. 2012; 5:493–503. PMID: 22705769.25. Fernandez-Sada E, Silva-Platas C, Villegas CA, Rivero SL, Willis BC, Garcia N, Garza JR, Oropeza-Almazan Y, Valverde CA, Mazzocchi G, Zazueta C, Torre-Amione G, Garcia-Rivas G. Cardiac responses to β-adrenoceptor stimulation is partly dependent on mitochondrial calcium uniporter activity. Br J Pharmacol. 2014; 171:4207–4221. PMID: 24628066.
Article26. Cook GA, Lavrentyev EN, Pham K, Park EA. Streptozotocin diabetes increases mRNA expression of ketogenic enzymes in the rat heart. Biochim Biophys Acta. 2017; 1861:307–312. PMID: 27845231.
Article27. Battiprolu PK, Rodnick KJ. Dichloroacetate selectively improves cardiac function and metabolism in female and male rainbow trout. Am J Physiol Heart Circ Physiol. 2014; 307:H1401–H1411. PMID: 25217653.
Article28. Le Page LM, Rider OJ, Lewis AJ, Ball V, Clarke K, Johansson E, Carr CA, Heather LC, Tyler DJ. Increasing pyruvate dehydrogenase flux as a treatment for diabetic cardiomyopathy: a combined 13C hyperpolarized magnetic resonance and echocardiography study. Diabetes. 2015; 64:2735–2743. PMID: 25795215.
Article29. Crewe C, Schafer C, Lee I, Kinter M, Szweda LI. Regulation of pyruvate dehydrogenase kinase 4 in the heart through degradation by the Lon protease in response to mitochondrial substrate availability. J Biol Chem. 2017; 292:305–312. PMID: 27856638.
Article30. Nadtochiy SM, Urciuoli W, Zhang J, Schafer X, Munger J, Brookes PS. Metabolomic profiling of the heart during acute ischemic preconditioning reveals a role for SIRT1 in rapid cardioprotective metabolic adaptation. J Mol Cell Cardiol. 2015; 88:64–72. PMID: 26388263.
Article31. Peters SJ, Harris RA, Wu P, Pehleman TL, Heigenhauser GJ, Spriet LL. Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am J Physiol Endocrinol Metab. 2001; 281:E1151–E1158. PMID: 11701428.32. DeFronzo RA, Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009; 32(Suppl 2):S157–S163. PMID: 19875544.
Article33. Guo Z. Pyruvate dehydrogenase, Randle cycle, and skeletal muscle insulin resistance. Proc Natl Acad Sci U S A. 2015; 112:E2854. PMID: 25941416.
Article34. Herbst EA, MacPherson RE, LeBlanc PJ, Roy BD, Jeoung NH, Harris RA, Peters SJ. Pyruvate dehydrogenase kinase-4 contributes to the recirculation of gluconeogenic precursors during postexercise glycogen recovery. Am J Physiol Regul Integr Comp Physiol. 2014; 306:R102–R107. PMID: 24305065.
Article35. Constantin-Teodosiu D, Stephens FB, Greenhaff PL. Perpetual muscle PDH activation in PDH kinase knockout mice protects against high-fat feeding-induced muscle insulin resistance. Proc Natl Acad Sci U S A. 2015; 112:E824. PMID: 25617371.
Article36. McAinch AJ, Cornall LM, Watts R, Hryciw DH, O'Brien PE, Cameron-Smith D. Increased pyruvate dehydrogenase kinase expression in cultured myotubes from obese and diabetic individuals. Eur J Nutr. 2015; 54:1033–1043. PMID: 25311062.
Article37. Cerutti R, Pirinen E, Lamperti C, Marchet S, Sauve AA, Li W, Leoni V, Schon EA, Dantzer F, Auwerx J, Viscomi C, Zeviani M. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 2014; 19:1042–1049. PMID: 24814483.
Article38. Tamaki M, Miyashita K, Wakino S, Mitsuishi M, Hayashi K, Itoh H. Chronic kidney disease reduces muscle mitochondria and exercise endurance and its exacerbation by dietary protein through inactivation of pyruvate dehydrogenase. Kidney Int. 2014; 85:1330–1339. PMID: 24284514.
Article39. Palamiuc L, Schlagowski A, Ngo ST, Vernay A, Dirrig-Grosch S, Henriques A, Boutillier AL, Zoll J, Echaniz-Laguna A, Loeffler JP, Rene F. A metabolic switch toward lipid use in glycolytic muscle is an early pathologic event in a mouse model of amyotrophic lateral sclerosis. EMBO Mol Med. 2015; 7:526–546. PMID: 25820275.
Article40. Zhang D, Li Y, Yao X, Wang H, Zhao L, Jiang H, Yao X, Zhang S, Ye C, Liu W, Cao H, Yu S, Wang YC, Li Q, Jiang J, Liu Y, Zhang L, Liu Y, Iwai N, Wang H, Li J, Li J, Li X, Jin ZB, Ying H. MiR-182 regulates metabolic homeostasis by modulating glucose utilization in muscle. Cell Rep. 2016; 16:757–768. PMID: 27396327.
Article41. Consitt LA, Saxena G, Saneda A, Houmard JA. Age-related impairments in skeletal muscle PDH phosphorylation and plasma lactate are indicative of metabolic inflexibility and the effects of exercise training. Am J Physiol Endocrinol Metab. 2016; 311:E145–E156. PMID: 27221120.
Article42. Cox PJ, Kirk T, Ashmore T, Willerton K, Evans R, Smith A, Murray AJ, Stubbs B, West J, McLure SW, King MT, Dodd MS, Holloway C, Neubauer S, Drawer S, Veech RL, Griffin JL, Clarke K. Nutritional ketosis alters fuel preference and thereby endurance performance in athletes. Cell Metab. 2016; 24:256–268. PMID: 27475046.
Article43. Mahmood S, Birkaya B, Rideout TC, Patel MS. Lack of mitochondria-generated acetyl-CoA by pyruvate dehydrogenase complex downregulates gene expression in the hepatic de novo lipogenic pathway. Am J Physiol Endocrinol Metab. 2016; 311:E117–E127. PMID: 27166281.
Article44. Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014; 159:1615–1625. PMID: 25525879.
Article45. Tao R, Xiong X, Harris RA, White MF, Dong XC. Genetic inactivation of pyruvate dehydrogenase kinases improves hepatic insulin resistance induced diabetes. PLoS One. 2013; 8:e71997. PMID: 23940800.
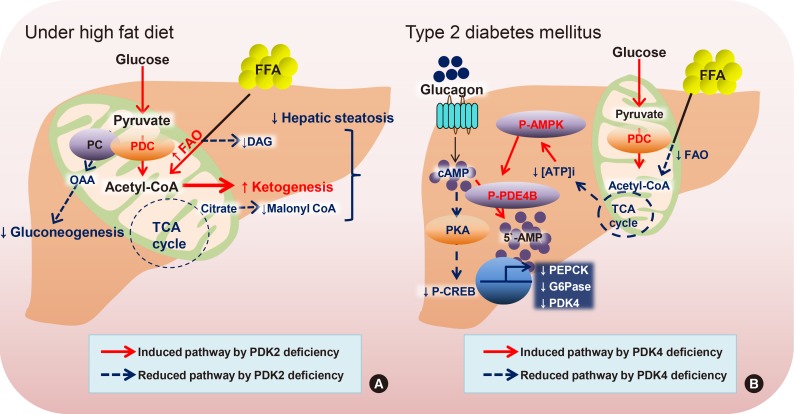
Article46. Go Y, Jeong JY, Jeoung NH, Jeon JH, Park BY, Kang HJ, Ha CM, Choi YK, Lee SJ, Ham HJ, Kim BG, Park KG, Park SY, Lee CH, Choi CS, Park TS, Lee WN, Harris RA, Lee IK. Inhibition of pyruvate dehydrogenase kinase 2 protects against hepatic steatosis through modulation of tricarboxylic acid cycle anaplerosis and ketogenesis. Diabetes. 2016; 65:2876–2887. PMID: 27385159.
Article47. Tso SC, Qi X, Gui WJ, Wu CY, Chuang JL, Wernstedt-Asterholm I, Morlock LK, Owens KR, Scherer PE, Williams NS, Tambar UK, Wynn RM, Chuang DT. Structure-guided development of specific pyruvate dehydrogenase kinase inhibitors targeting the ATP-binding pocket. J Biol Chem. 2014; 289:4432–4443. PMID: 24356970.
Article48. Jha MK, Jeon S, Suk K. Pyruvate dehydrogenase kinases in the nervous system: their principal functions in neuronal-glial metabolic interaction and neuro-metabolic disorders. Curr Neuropharmacol. 2012; 10:393–403. PMID: 23730261.49. Yao J, Chen S, Mao Z, Cadenas E, Brinton RD. 2-Deoxy-D-glucose treatment induces ketogenesis, sustains mitochondrial function, and reduces pathology in female mouse model of Alzheimer's disease. PLoS One. 2011; 6:e21788. PMID: 21747957.
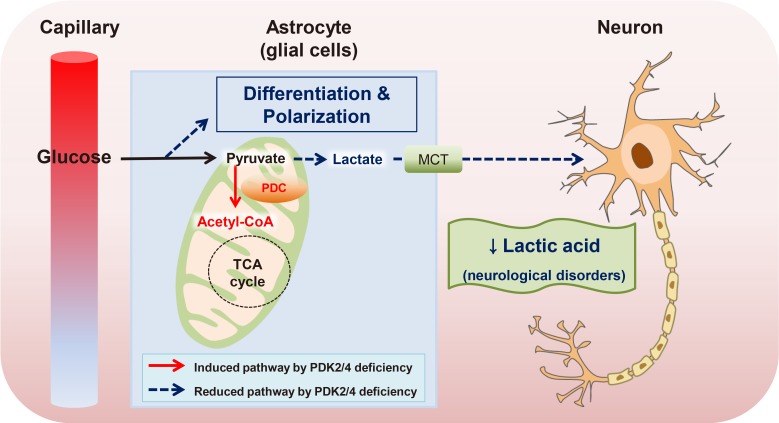
Article50. Geng X, Elmadhoun O, Peng C, Ji X, Hafeez A, Liu Z, Du H, Rafols JA, Ding Y. Ethanol and normobaric oxygen: novel approach in modulating pyruvate dehydrogenase complex after severe transient and permanent ischemic stroke. Stroke. 2015; 46:492–499. PMID: 25563647.51. Halim ND, Mcfate T, Mohyeldin A, Okagaki P, Korotchkina LG, Patel MS, Jeoung NH, Harris RA, Schell MJ, Verma A. Phosphorylation status of pyruvate dehydrogenase distinguishes metabolic phenotypes of cultured rat brain astrocytes and neurons. Glia. 2010; 58:1168–1176. PMID: 20544852.
Article52. Jha MK, Lee IK, Suk K. Metabolic reprogramming by the pyruvate dehydrogenase kinase-lactic acid axis: linking metabolism and diverse neuropathophysiologies. Neurosci Biobehav Rev. 2016; 68:1–19. PMID: 27179453.
Article53. Pliss L, Jatania U, Patel MS. Beneficial effect of feeding a ketogenic diet to mothers on brain development in their progeny with a murine model of pyruvate dehydrogenase complex deficiency. Mol Genet Metab Rep. 2016; 7:78–86. PMID: 27331005.
Article54. Asencio C, Rodriguez-Hernandez MA, Briones P, Montoya J, Cortes A, Emperador S, Gavilan A, Ruiz-Pesini E, Yubero D, Montero R, Pineda M, O'Callaghan MM, Alcazar-Fabra M, Salviati L, Artuch R, Navas P. Severe encephalopathy associated to pyruvate dehydrogenase mutations and unbalanced coenzyme Q10 content. Eur J Hum Genet. 2016; 24:367–372. PMID: 26014431.
Article55. Perez-Siles G, Ly C, Grant A, Drew AP, Yiu EM, Ryan MM, Chuang DT, Tso SC, Nicholson GA, Kennerson ML. Pathogenic mechanisms underlying X-linked Charcot-Marie-Tooth neuropathy (CMTX6) in patients with a pyruvate dehydrogenase kinase 3 mutation. Neurobiol Dis. 2016; 94:237–244. PMID: 27388934.
Article56. Fluge O, Mella O, Bruland O, Risa K, Dyrstad SE, Alme K, Rekeland IG, Sapkota D, Rosland GV, Fossa A, Ktoridou-Valen I, Lunde S, Sorland K, Lien K, Herder I, Thurmer H, Gotaas ME, Baranowska KA, Bohnen LM, Schafer C, McCann A, Sommerfelt K, Helgeland L, Ueland PM, Dahl O, Tronstad KJ. Metabolic profiling indicates impaired pyruvate dehydrogenase function in myalgic encephalopathy/chronic fatigue syndrome. JCI Insight. 2016; 1:e89376. PMID: 28018972.
Article57. Dugan LL, You YH, Ali SS, Diamond-Stanic M, Miyamoto S, DeCleves AE, Andreyev A, Quach T, Ly S, Shekhtman G, Nguyen W, Chepetan A, Le TP, Wang L, Xu M, Paik KP, Fogo A, Viollet B, Murphy A, Brosius F, Naviaux RK, Sharma K. AMPK dysregulation promotes diabetes-related reduction of superoxide and mitochondrial function. J Clin Invest. 2013; 123:4888–4899. PMID: 24135141.
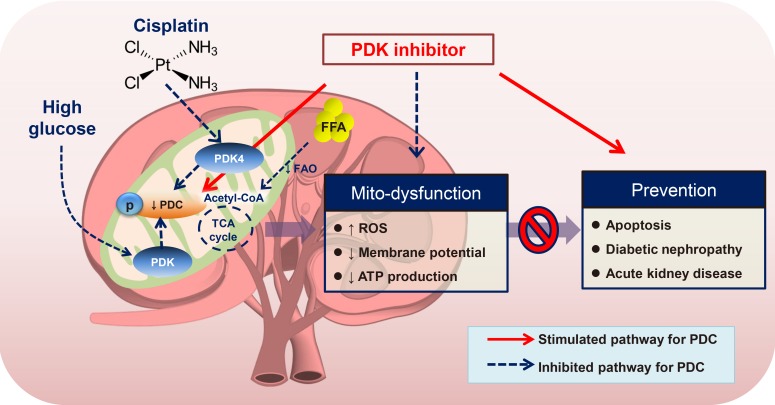
Article58. Oh CJ, Ha CM, Choi YK, Park S, Choe MS, Jeoung NH, Huh YH, Kim HJ, Kweon HS, Lee JM, Lee SJ, Jeon JH, Harris RA, Park KG, Lee IK. Pyruvate dehydrogenase kinase 4 deficiency attenuates cisplatin-induced acute kidney injury. Kidney Int. 2017; 91:880–895. PMID: 28040265.
Article59. Lee H, Abe Y, Lee I, Shrivastav S, Crusan AP, Huttemann M, Hopfer U, Felder RA, Asico LD, Armando I, Jose PA, Kopp JB. Increased mitochondrial activity in renal proximal tubule cells from young spontaneously hypertensive rats. Kidney Int. 2014; 85:561–569. PMID: 24132210.
Article60. Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, Zhang H, Lin C, Qi NR, Michailidis G, Groop PH, Nelson RG, Darshi M, Sharma K, Schelling JR, Sedor JR, Pop-Busui R, Weinberg JM, Soleimanpour SA, Abcouwer SF, Gardner TW, Burant CF, Feldman EL, Kretzler M, Brosius FC 3rd, Pennathur S. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 2016; 1:e86976. PMID: 27699244.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Role of Pyruvate Dehydrogenase Kinase in Diabetes and Obesity
- Pyruvate Dehydrogenase Kinases: Therapeutic Targets for Diabetes and Cancers
- Anesthetic experience in a pediatric patient with pyruvate dehydrogenase complex (PDHC) deficiency: A case report
- Regulation of mammalian pyruvate dehydrogenase complex by phosphorylation: complexity of multiple phosphorylation sites and kinases
- The Link between Mitochondrial Dysfunction and Sarcopenia: An Update Focusing on the Role of Pyruvate Dehydrogenase Kinase 4