J Breast Cancer.
2018 Dec;21(4):371-381. 10.4048/jbc.2018.21.e60.
miR-195/miR-497 Regulate CD274 Expression of Immune Regulatory Ligands in Triple-Negative Breast Cancer
- Affiliations
-
- 1The 1st Ward of the Medical Department, Affiliated Cancer Hospital and Institute of Guangzhou Medical University, Guangzhou, China. chhpan@163.com
- 2Radiotherapy Department, Central Hospital of Guangdong Nongken, Zhanjiang, China.
- 3State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, China.
- KMID: 2429815
- DOI: http://doi.org/10.4048/jbc.2018.21.e60
Abstract
- PURPOSE
Immune suppression is common in patients with advanced breast cancer but the mechanisms underlying this phenomenon have not been sufficiently studied. In this study, we aimed to identify B7 family members that were able to predict the immune status of patients, and which may serve as potential targets for the treatment of breast cancer. We also aimed to identify microRNAs that may regulate the expression of B7 family members.
METHODS
The Cancer Genome Atlas data from 1,092 patients with breast cancer, including gene expression, microRNA expression and survival data, were used for statistical and survival analyses. Polymerase chain reaction and Western blot were used to measure messenger RNA and protein expression, respectively. Luciferase assay was used to investigate direct microRNA target.
RESULTS
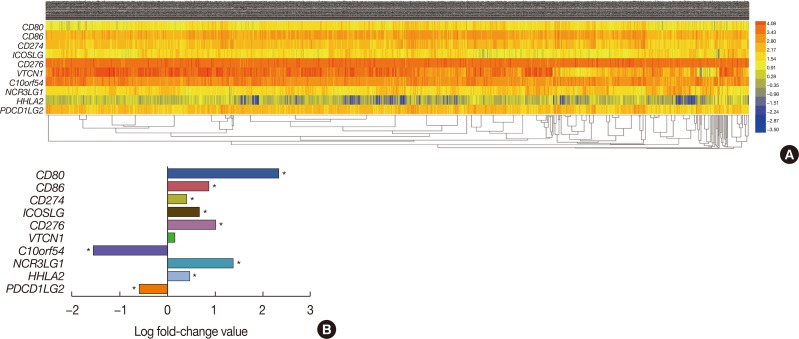
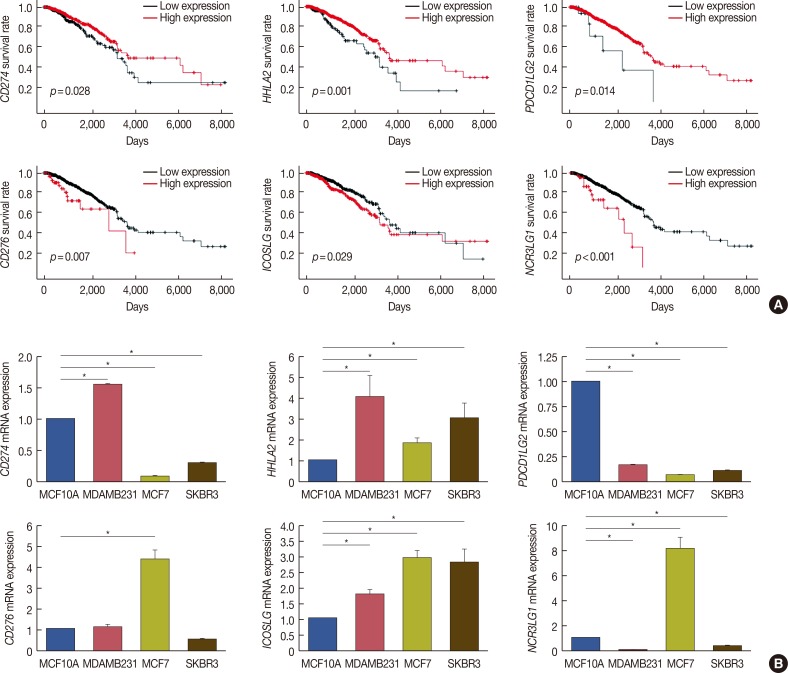
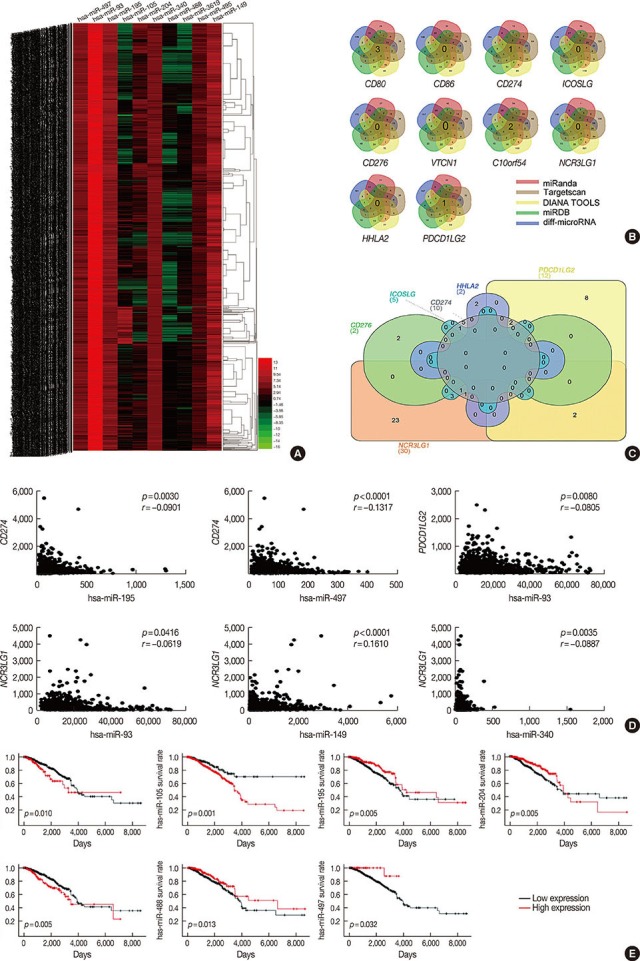
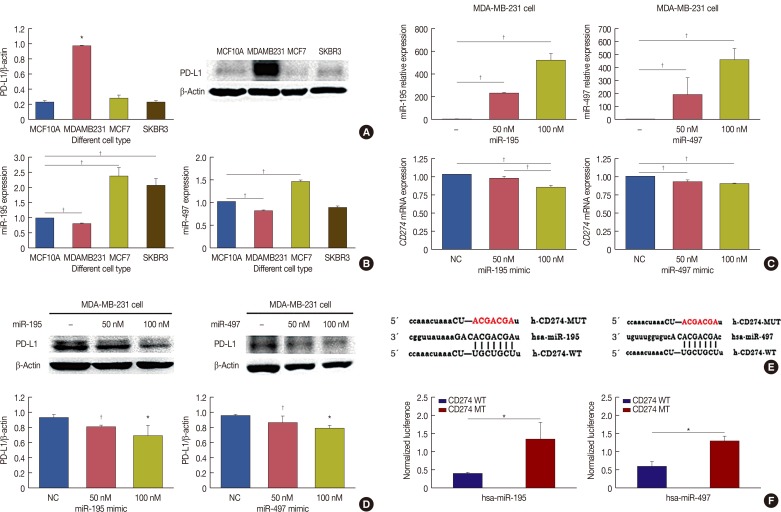
Bioinformatic analysis predicted that microRNA (miR)-93, miR-195, miR-497, and miR-340 are potential regulators of the immune evasion of breast cancer cells, and that they exert this function by targeting CD274, PDCD1LG2, and NCR3LG1. We chose CD274 for further investigations. We found that miR-195, miR-497, and CD274 expression levels were inversely correlated in MDA-MB-231 cells, and miR-195 and miR-497 expressions mimic inhibited CD274 expression in vitro. Mechanistic investigations demonstrated that miR-195 and miR-497 directly target CD274 3"² untranslated region.
CONCLUSION
Our data indicated that the level of B7 family members can predict the prognosis of breast cancer patients, and miR-195/miR-497 regulate CD274 expression in triple negative breast cancer. This regulation may further influence tumor progression and the immune tolerance mechanism in breast cancer and may be able to predict the effect of immunotherapy on patients.
Keyword
MeSH Terms
-
Antigens, CD274
B7 Antigens
Blotting, Western
Breast Neoplasms
Computational Biology
Gene Expression
Genome
Humans
Immune Evasion
Immune Tolerance
Immunotherapy
In Vitro Techniques
Ligands*
Luciferases
MicroRNAs
Polymerase Chain Reaction
Prognosis
RNA, Messenger
Triple Negative Breast Neoplasms*
Untranslated Regions
Antigens, CD274
B7 Antigens
Ligands
Luciferases
MicroRNAs
RNA, Messenger
Untranslated Regions
Figure
Reference
-
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–E386. PMID: 25220842.
Article2. DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016; 66:31–42. PMID: 26513636.
Article3. Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol. 2014; 15:e279–e289. PMID: 24872111.
Article4. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015; 15:321–333. PMID: 25998712.
Article5. Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008; 8:467–477. PMID: 18500231.
Article6. Schildberg FA, Klein SR, Freeman GJ, Sharpe AH. Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity. 2016; 44:955–972. PMID: 27192563.
Article7. Callahan MK, Postow MA, Wolchok JD. Targeting T cell co-receptors for cancer therapy. Immunity. 2016; 44:1069–1078. PMID: 27192570.
Article8. Zhao L, Yu H, Yi S, Peng X, Su P, Xiao Z, et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016; 7:45370–45384. PMID: 27248318.
Article9. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat Commun. 2016; 7:11406. PMID: 27147225.
Article10. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. PMID: 19910308.
Article11. Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012; 7:e51862. PMID: 23251644.
Article12. Gibson J. Anti-PD-L1 for metastatic triple-negative breast cancer. Lancet Oncol. 2015; 16:e264.
Article13. Wu R, Li F, Zhu J, Tang R, Qi Q, Zhou X, et al. A functional variant at miR-132-3p, miR-212-3p, and miR-361-5p binding site in CD80 gene alters susceptibility to gastric cancer in a Chinese Han population. Med Oncol. 2014; 31:60. PMID: 24981235.
Article14. Yuan SM, Li H, Yang M, Zha H, Sun H, Li XR, et al. High intensity focused ultrasound enhances anti-tumor immunity by inhibiting the negative regulatory effect of miR-134 on CD86 in a murine melanoma model. Oncotarget. 2015; 6:37626–37637. PMID: 26485753.
Article15. Wu L, Chen Z, Zhang J, Xing Y. Effect of miR-513a-5p on etoposide-stimulating B7-H1 expression in retinoblastoma cells. J Huazhong Univ Sci Technolog Med Sci. 2012; 32:601–606. PMID: 22886978.
Article16. Guo W, Tan W, Liu S, Huang X, Lin J, Liang R, et al. MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumour Biol. 2015; 36:9049–9057. PMID: 26084609.
Article17. Zhu J, Chen L, Zou L, Yang P, Wu R, Mao Y, et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol. 2014; 75:348–353. PMID: 24468585.
Article18. Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014; 5:5241. PMID: 25348003.
Article19. Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015; 27:443–452. PMID: 25499621.
Article20. Yang P, Tang R, Zhu J, Zou L, Wu R, Zhou H, et al. A functional variant at miR-24 binding site in B7-H2 alters susceptibility to gastric cancer in a Chinese Han population. Mol Immunol. 2013; 56:98–103. PMID: 23688438.
Article21. Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009; 69:6275–6281. PMID: 19584290.
Article22. Wang ZS, Zhong M, Bian YH, Mu YF, Qin SL, Yu MH, et al. MicroRNA-187 inhibits tumor growth and invasion by directly targeting CD276 in colorectal cancer. Oncotarget. 2016; 7:44266–44276. PMID: 27329595.
Article23. Wang L, Kang FB, Sun N, Wang J, Chen W, Li D, et al. The tumor suppressor miR-124 inhibits cell proliferation and invasion by targeting B7-H3 in osteosarcoma. Tumour Biol. 2016; 37:14939–14947. PMID: 27644254.
Article24. Zhou X, Mao Y, Zhu J, Meng F, Chen Q, Tao L, et al. TGF-beta1 promotes colorectal cancer immune escape by elevating B7-H3 and B7-H4 via the miR-155/miR-143 axis. Oncotarget. 2016; 7:67196–67211. PMID: 27626488.25. Muenst S, Schaerli AR, Gao F, Däster S, Trella E, Droeser RA, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2014; 146:15–24. PMID: 24842267.
Article26. Baptista MZ, Sarian LO, Derchain SF, Pinto GA, Vassallo J. Prognostic significance of PD-L1 and PD-L2 in breast cancer. Hum Pathol. 2016; 47:78–84. PMID: 26541326.
Article27. Faget J, Sisirak V, Blay JY, Caux C, Bendriss-Vermare N, Ménétrier-Caux C. ICOS is associated with poor prognosis in breast cancer as it promotes the amplification of immunosuppressive CD4(+) T cells by plasmacytoid dendritic cells. Oncoimmunology. 2013; 2:e23185. PMID: 23802069.28. Mao Y, Li W, Chen K, Xie Y, Liu Q, Yao M, et al. B7-H1 and B7-H3 are independent predictors of poor prognosis in patients with non-small cell lung cancer. Oncotarget. 2015; 6:3452–3461. PMID: 25609202.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple Negative Breast Cancer
- LncRNA DLG1-AS1 Promotes Cancer Cell Proliferation in Triple Negative Breast Cancer by Downregulating miR-203
- The Significance of MicroRNA Let-7b, miR-30c, and miR-200c Expression in Breast Cancers
- Low Expression of Circulating MicroRNA-34c is Associated with Poor Prognosis in Triple-Negative Breast Cancer
- MicroRNA-222 Expression as a Predictive Marker for Tumor Progression in Hormone Receptor-Positive Breast Cancer