Cancer Res Treat.
2016 Jan;48(1):322-333. 10.4143/crt.2014.294.
Interactome Analysis Reveals that Heterochromatin Protein 1gamma (HP1gamma) Is Associated with the DNA Damage Response Pathway
- Affiliations
-
- 1Department of Biological Sciences, Sungkyunkwan University, Suwon, Korea.
- 2Genomic Instability Research Center, Ajou University School of Medicine, Suwon, Korea. hckang@ajou.ac.kr, jsjlee@ajou.ac.kr
- 3Department of Life Sciences, College of Natural Sciences, Ajou University, Suwon, Korea.
- 4Leading-edge Research Center for Drug Discovery and Development and Metabolic Disease, Kyungpook National University, Daegu, Korea.
- KMID: 2152291
- DOI: http://doi.org/10.4143/crt.2014.294
Abstract
- PURPOSE
Heterochromatin protein 1gamma (HP1gamma) interacts with chromosomes by binding to lysine 9-methylated histone H3 or DNA/RNA. HP1gamma is involved in various biological processes. The purpose of this study is to gain an understanding of how HP1gamma functions in these processes by identifying HP1gamma-binding proteins using mass spectrometry.
MATERIALS AND METHODS
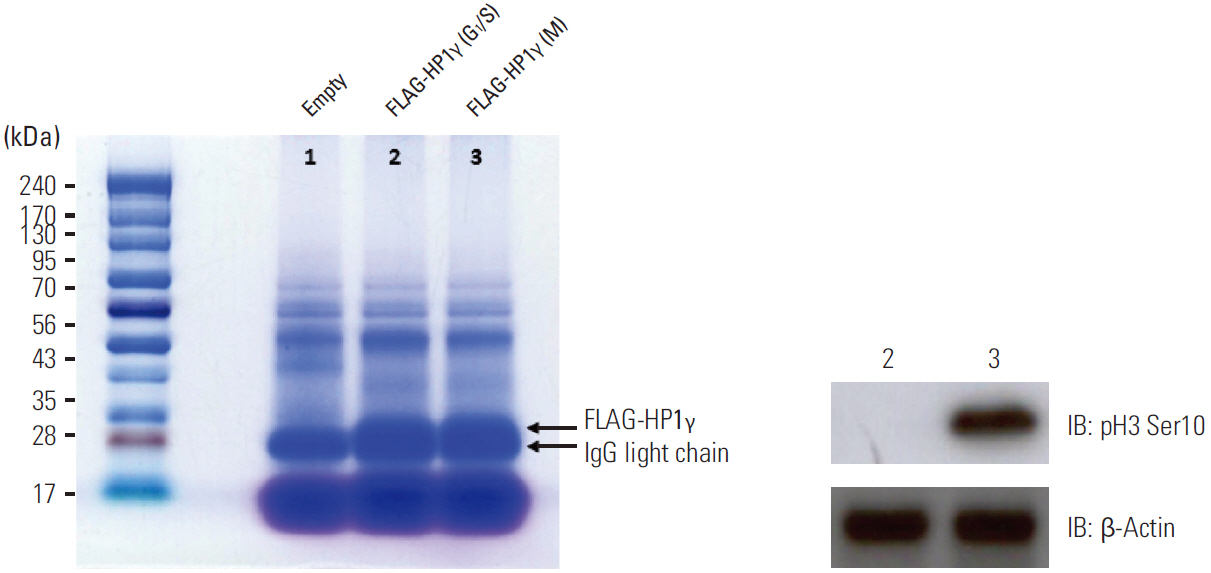
We performed affinity purification of HP1gamma-binding proteins using G1/S phase or prometaphase HEK293T cell lysates that transiently express mock or FLAG-HP1gamma. Coomassie staining was performed for HP1gamma-binding complexes, using cell lysates prepared by affinity chromatography FLAG-agarose beads, and the bands were digested and then analyzed using a mass spectrometry.
RESULTS
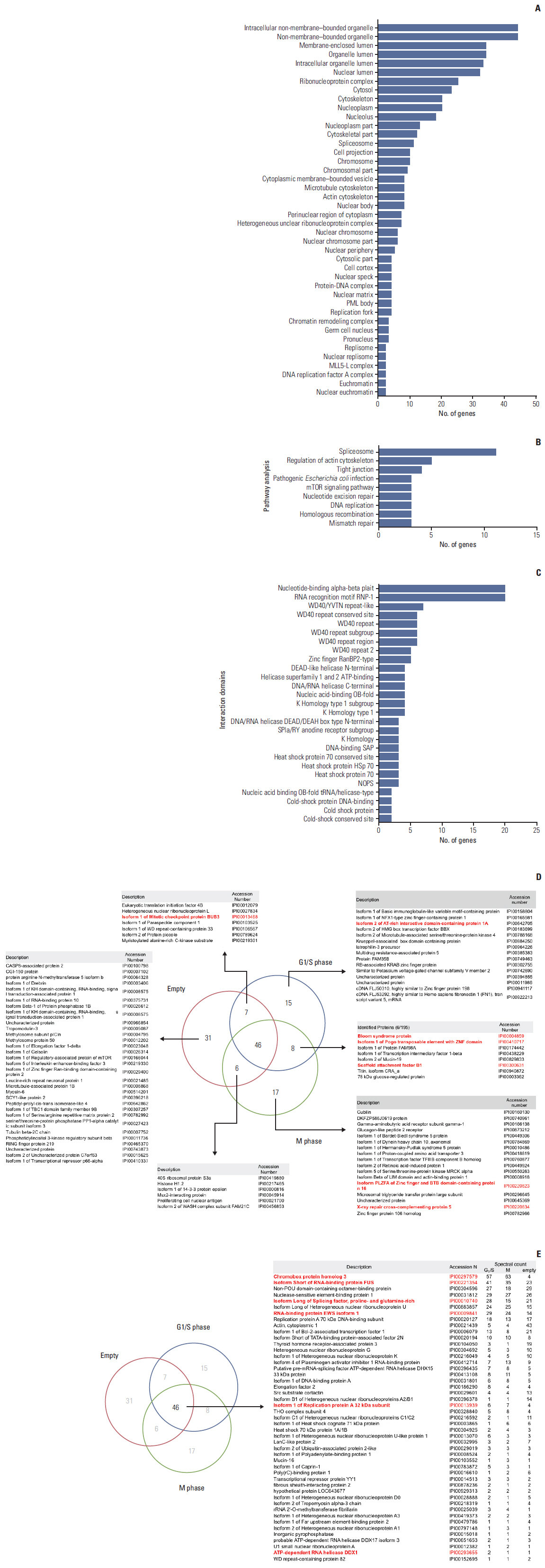
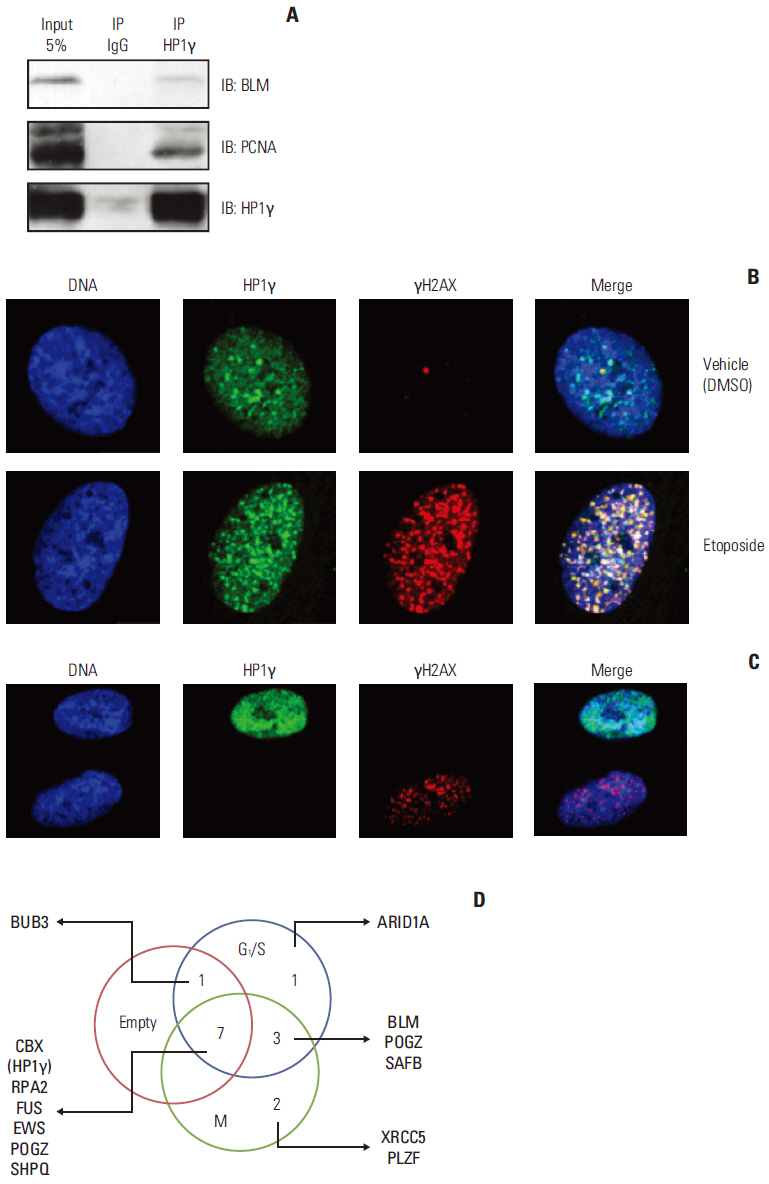
We identified 99 HP1gamma-binding proteins with diverse cellular functions, including spliceosome, regulation of the actin cytoskeleton, tight junction, pathogenic Escherichia coli infection, mammalian target of rapamycin signaling pathway, nucleotide excision repair, DNA replication, homologous recombination, and mismatch repair.
CONCLUSION
Our results suggested that HP1gamma is functionally active in DNA damage response via protein-protein interaction.
Keyword
MeSH Terms
-
Actin Cytoskeleton
Biological Processes
Chromatography, Affinity
DNA Damage*
DNA Mismatch Repair
DNA Repair
DNA Replication
DNA*
Escherichia coli Infections
Heterochromatin*
Histones
Homologous Recombination
Lysine
Mass Spectrometry
Prometaphase
Sirolimus
Spliceosomes
Tight Junctions
DNA
Heterochromatin
Histones
Lysine
Sirolimus
Figure
Reference
-
References
1. Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004; 5:296–304.
Article2. Jacobs SA, Khorasanizadeh S. Struc ture of H P1 chr omodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002; 295:2080–3.3. Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000; 10:204–10.
Article4. Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000; 90:279–84.5. Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005; 19:381–91.6. du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007; 26:424–35.7. El Gazzar M, Yoza BK, Chen X, Hu J, Hawkins GA, McCall CE. G9a and HP1 couple histone and DNA methylation to TNFalpha transcription silencing during endotoxin tolerance. J Biol Chem. 2008; 283:32198–208.8. Abe K, Naruse C, Kato T, Nishiuchi T, Saitou M, Asano M. Loss of heterochromatin protein 1 gamma reduces the number of primordial germ cells via impaired cell cycle progression in mice. Biol Reprod. 2011; 85:1013–24.9. Liu X, Song Z, Huo Y, Zhang J, Zhu T, Wang J, et al. Chromatin protein HP1 interacts with the mitotic regulator borealin protein and specifies the centromere localization of the chromosomal passenger complex. J Biol Chem. 2014; 289:20638–49.
Article10. Choi JD, Park MA, Lee JS. Suppression and recovery of BRCA1-mediated transcription by HP1gamma via modulation of promoter occupancy. Nucleic Acids Res. 2012; 40:11321–38.11. Cho HJ, Oh YJ, Han SH, Chung HJ, Kim CH, Lee NS, et al. Cdk1 protein-mediated phosphorylation of receptor-associated protein 80 (RAP80) serine 677 modulates DNA damage-induced G2/M checkpoint and cell survival. J Biol Chem. 2013; 288:3768–76.
Article12. Cho HJ, Lee EH, Han SH, Chung HJ, Jeong JH, Kwon J, et al. Degradation of human RAP80 is cell cycle regulated by Cdc20 and Cdh1 ubiquitin ligases. Mol Cancer Res. 2012; 10:615–25.
Article13. Li L, Monckton EA, Godbout R. A role for DEAD box 1 at DNA double-strand breaks. Mol Cell Biol. 2008; 28:6413–25.
Article14. Nozawa RS, Nagao K, Masuda HT, Iwasaki O, Hirota T, Nozaki N, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010; 12:719–27.15. Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003; 112:1751–61.
Article16. Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013; 16:1383–91.
Article17. Paronetto MP. Ewing sarcoma protein: a key player in human cancer. Int J Cell Biol. 2013; 2013:642853.
Article18. Lee YH, Kuo CY, Stark JM, Shih HM, Ann DK. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 2013; 41:5784–98.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Heterochromatin-1 Phosphorylation Contributes to TPA-Induced AP-1 Expression
- Mad2B forms a complex with Cdc20, Cdc27, Rev3 and Rev1 in response to cisplatin-induced DNA damage
- Inactivation of Mad2B Enhances Apoptosis in Human Cervical Cancer Cell Line upon Cisplatin-Induced DNA Damage
- Analysis of Ultraviolet Light Damage in Mammalian Cells by Flowcytometry
- Biological Network Evolution Hypothesis Applied to Protein Structural Interactome