Endocrinol Metab.
2014 Dec;29(4):435-440. 10.3803/EnM.2014.29.4.435.
Roles of Protein Arginine Methyltransferases in the Control of Glucose Metabolism
- Affiliations
-
- 1Department of Life Sciences, Korea University College of Life Sciences and Biotechnology, Seoul, Korea. koohoi@korea.ac.kr
- KMID: 2384238
- DOI: http://doi.org/10.3803/EnM.2014.29.4.435
Abstract
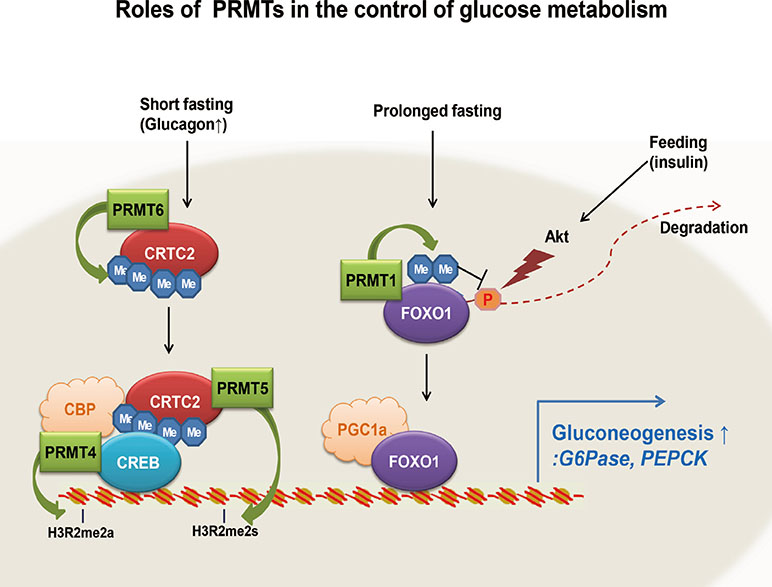
- Glucose homeostasis is tightly controlled by the regulation of glucose production in the liver and glucose uptake into peripheral tissues, such as skeletal muscle and adipose tissue. Under prolonged fasting, hepatic gluconeogenesis is mainly responsible for glucose production in the liver, which is essential for tissues, organs, and cells, such as skeletal muscle, the brain, and red blood cells. Hepatic gluconeogenesis is controlled in part by the concerted actions of transcriptional regulators. Fasting signals are relayed by various intracellular enzymes, such as kinases, phosphatases, acetyltransferases, and deacetylases, which affect the transcriptional activity of transcription factors and transcriptional coactivators for gluconeogenic genes. Protein arginine methyltransferases (PRMTs) were recently added to the list of enzymes that are critical for regulating transcription in hepatic gluconeogenesis. In this review, we briefly discuss general aspects of PRMTs in the control of transcription. More specifically, we summarize the roles of four PRMTs: PRMT1, PRMT 4, PRMT 5, and PRMT 6, in the control of hepatic gluconeogenesis through specific regulation of FoxO1- and CREB-dependent transcriptional events.
MeSH Terms
-
Acetyltransferases
Adipose Tissue
Arginine*
Brain
Erythrocytes
Fasting
Gluconeogenesis
Glucose*
Homeostasis
Liver
Metabolism*
Methyltransferases*
Muscle, Skeletal
Phosphoric Monoester Hydrolases
Phosphotransferases
Protein-Arginine N-Methyltransferases
Transcription Factors
Acetyltransferases
Arginine
Glucose
Methyltransferases
Phosphoric Monoester Hydrolases
Phosphotransferases
Protein-Arginine N-Methyltransferases
Transcription Factors
Figure
Cited by 1 articles
-
Protein Arginine Methyltransferases: Emerging Targets in Cardiovascular and Metabolic Disease
Yan Zhang, Shibo Wei, Eun-Ju Jin, Yunju Jo, Chang-Myung Oh, Gyu-Un Bae, Jong-Sun Kang, Dongryeol Ryu
Diabetes Metab J. 2024;48(4):487-502. doi: 10.4093/dmj.2023.0362.
Reference
-
1. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009; 33:1–13.2. Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013; 13:37–50.3. Feng Y, Wang J, Asher S, Hoang L, Guardiani C, Ivanov I, Zheng YG. Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J Biol Chem. 2011; 286:20323–20334.4. Feng Q, Yi P, Wong J, O'Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006; 26:7846–7857.5. Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007; 25:71–83.6. Lacroix M, El Messaoudi S, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008; 9:452–458.7. Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007; 27:384–394.8. Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem J. 2005; 388(Pt 1):379–386.9. Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000; 275:7723–7730.10. Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, Allis CD, Tempst P, Nimer SD. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008; 22:640–653.11. Biggs WH 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999; 96:7421–7426.12. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999; 96:857–868.13. Nakae J, Kitamura T, Kitamura Y, Biggs WH 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003; 4:119–129.14. Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008; 32:221–231.15. Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003; 423:550–555.16. Ayala JE, Streeper RS, Desgrosellier JS, Durham SK, Suwanichkul A, Svitek CA, Goldman JK, Barr FG, Powell DR, O'Brien RM. Conservation of an insulin response unit between mouse and human glucose-6-phosphatase catalytic subunit gene promoters: transcription factor FKHR binds the insulin response sequence. Diabetes. 1999; 48:1885–1889.17. Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007; 6:208–216.18. Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006; 281:10105–10117.19. Choi D, Oh KJ, Han HS, Yoon YS, Jung CY, Kim ST, Koo SH. Protein arginine methyltransferase 1 regulates hepatic glucose production in a FoxO1-dependent manner. Hepatology. 2012; 56:1546–1556.20. Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999; 284:2174–2177.21. Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005; 18:263–272.22. Ceschin DG, Walia M, Wenk SS, Duboe C, Gaudon C, Xiao Y, Fauquier L, Sankar M, Vandel L, Gronemeyer H. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011; 25:1132–1146.23. Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002; 21:5457–5466.24. Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J Biol Chem. 2002; 277:4324–4333.25. Miao F, Li S, Chavez V, Lanting L, Natarajan R. Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol Endocrinol. 2006; 20:1562–1573.26. Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008; 9:193–198.27. Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, Zhang S, Li L, Pan CX, Hu Z, MacLennan GT, Cheng L. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004; 101:83–89.28. Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002; 277:44623–44630.29. Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001; 413:179–183.30. Erion DM, Ignatova ID, Yonemitsu S, Nagai Y, Chatterjee P, Weismann D, Hsiao JJ, Zhang D, Iwasaki T, Stark R, Flannery C, Kahn M, Carmean CM, Yu XX, Murray SF, Bhanot S, Monia BP, Cline GW, Samuel VT, Shulman GI. Prevention of hepatic steatosis and hepatic insulin resistance by knockdown of cAMP response element-binding protein. Cell Metab. 2009; 10:499–506.31. Krones-Herzig A, Mesaros A, Metzger D, Ziegler A, Lemke U, Bruning JC, Herzig S. Signal-dependent control of gluconeogenic key enzyme genes through coactivator-associated arginine methyltransferase 1. J Biol Chem. 2006; 281:3025–3029.32. Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001; 294:2507–2511.33. Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003; 23:7475–7487.34. Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ 3rd. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008; 28:3198–3207.35. Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006; 8:623–630.36. Tsai WW, Niessen S, Goebel N, Yates JR 3rd, Guccione E, Montminy M. PRMT5 modulates the metabolic response to fasting signals. Proc Natl Acad Sci USA. 2013; 110:8870–8875.37. Kanamaluru D, Xiao Z, Fang S, Choi SE, Kim DH, Veenstra TD, Kemper JK. Arginine methylation by PRMT5 at a naturally occurring mutation site is critical for liver metabolic regulation by small heterodimer partner. Mol Cell Biol. 2011; 31:1540–1550.38. Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007; 21:3369–3380.39. Harrison MJ, Tang YH, Dowhan DH. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 2010; 38:2201–2216.40. Han HS, Jung CY, Yoon YS, Choi S, Choi D, Kang G, Park KG, Kim ST, Koo SH. Arginine methylation of CRTC2 is critical in the transcriptional control of hepatic glucose metabolism. Sci Signal. 2014; 7:ra19.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Arginine methylation as a key post-translational modification in skeletal muscle homeostasis: a review
- Protein Arginine Methyltransferases: Emerging Targets in Cardiovascular and Metabolic Disease
- Identification of Protein Arginine Methyltransferase 5 as a Regulator for Encystation of Acanthamoeba
- Methylation of eukaryotic elongation factor 2 induced by basic fibroblast growth factor via mitogen-activated protein kinase
- Protein methylation in cellular proliferation and differentiation: Non-histone nuclear methyl acceptor protein(s) during 3'-methyl-4-dimethylaminoazobenzeneinduced hepatocarcinogenesis