J Korean Med Sci.
2012 Jun;27(6):594-604. 10.3346/jkms.2012.27.6.594.
Pharmacological Unmasking Microarray Approach-Based Discovery of Novel DNA Methylation Markers for Hepatocellular Carcinoma
- Affiliations
-
- 1Laboratory of Epigenetics, Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea. ghkang@snu.ac.kr
- 2Department of Pathology, Seoul National University College of Medicine, Seoul, Korea.
- 3Department of General Surgery, Seoul National University College of Medicine, Seoul, Korea.
- 4Department of Pathology, Korea University School of Medicine, Seoul, Korea.
- KMID: 1421610
- DOI: http://doi.org/10.3346/jkms.2012.27.6.594
Abstract
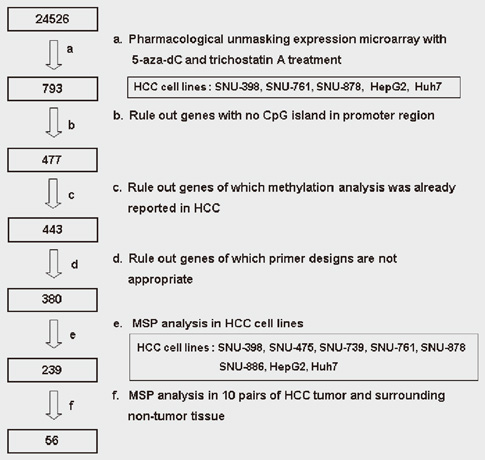
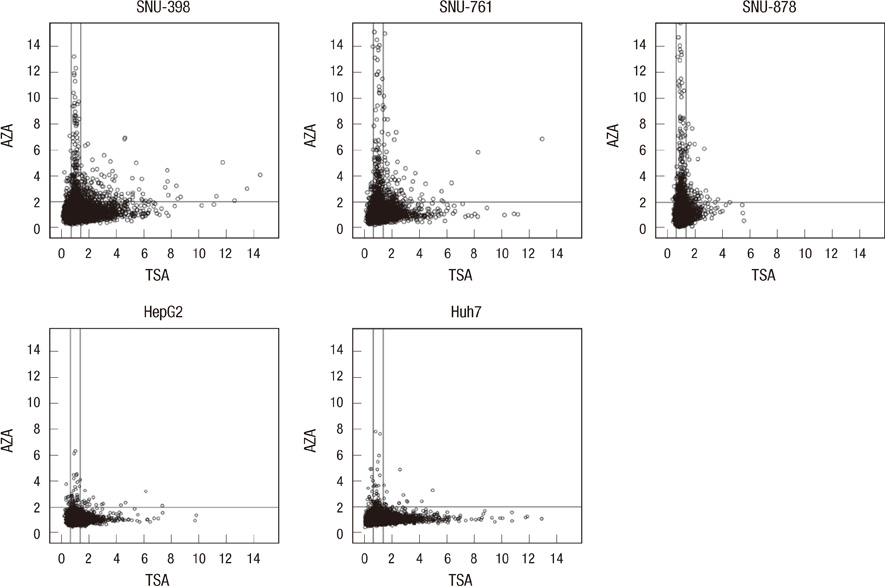
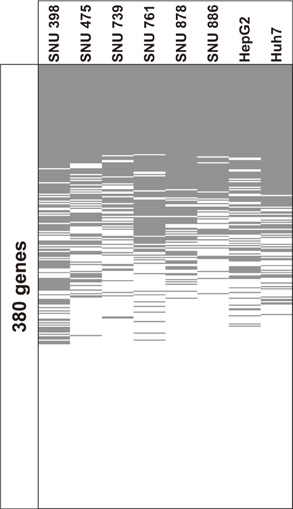
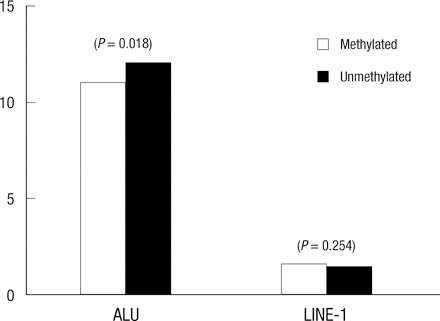
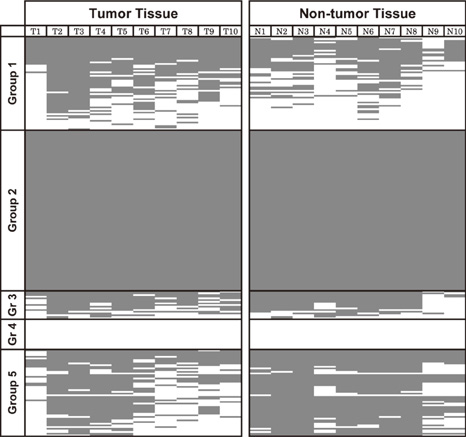
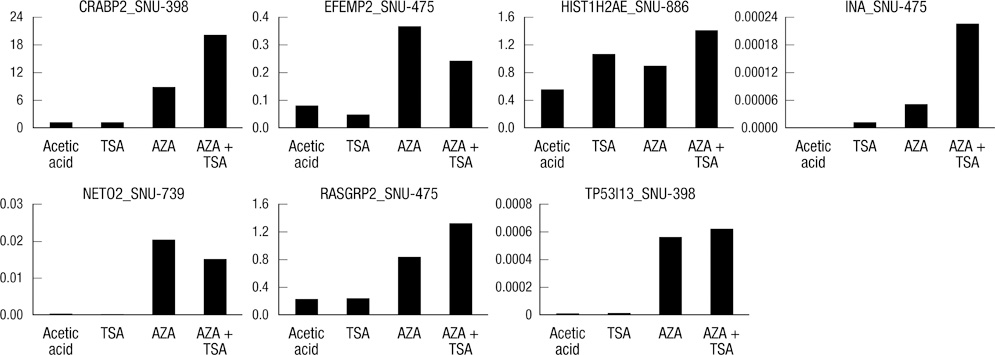
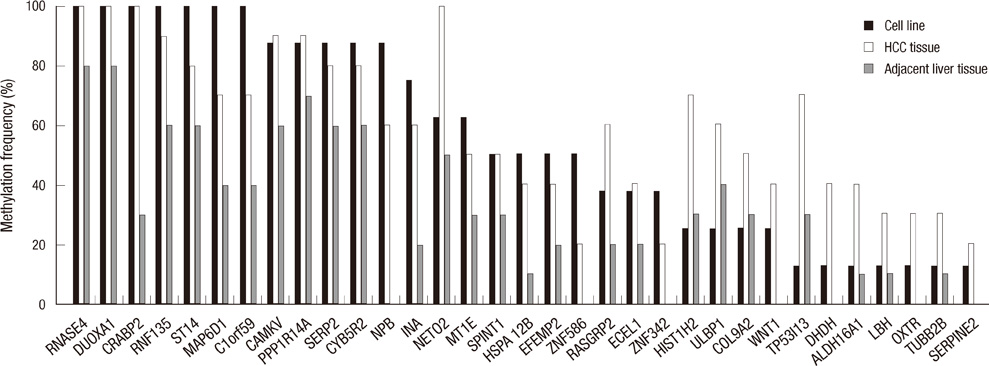
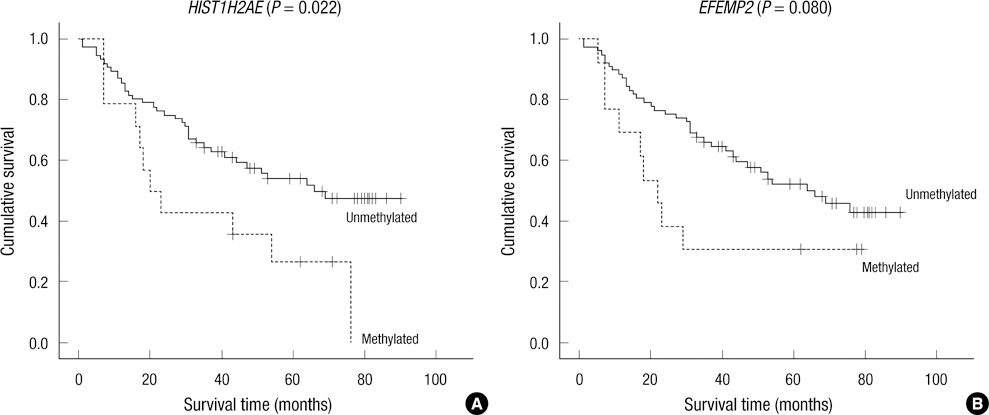
- DNA methylation is one of the main epigenetic mechanisms and hypermethylation of CpG islands at tumor suppressor genes switches off these genes. To find novel DNA methylation markers in hepatocellular carcinoma (HCC), we performed pharmacological unmasking (treatment with 5-aza-2'-deoxycytidine or trichostatin A) followed by microarray analysis in HCC cell lines. Of the 239 promoter CpG island loci hypermethylated in HCC cell lines (as revealed by methylation-specific PCR), 221 loci were found to be hypermethylated in HCC or nonneoplastic liver tissues. Thirty-three loci showed a 20% higher methylation frequency in tumors than in adjacent nonneoplastic tissues. Correlation of individual cancer-related methylation markers with clinicopathological features of HCC patients (n = 95) revealed that the number of hypermethylated genes in HCC tumors was higher in older than in younger patients. Univariate and multivariate survival analysis revealed that the HIST1H2AE methylation status is closely correlated with the patient's overall survival (P = 0.022 and P = 0.010, respectively). In conclusion, we identified 221 novel DNA methylation markers for HCC. One promising prognostic marker, HIST1H2AE, should be further validated in the prognostication of HCC patients.
MeSH Terms
-
Azacitidine/analogs & derivatives/pharmacology
Carcinoma, Hepatocellular/*genetics/mortality
Cell Line, Tumor
CpG Islands
DNA Methylation/*drug effects
Down-Regulation
Female
Hep G2 Cells
Humans
Hydroxamic Acids/pharmacology
Liver/metabolism
Liver Neoplasms/*genetics/mortality
Male
Middle Aged
Oligonucleotide Array Sequence Analysis
Promoter Regions, Genetic
Survival Analysis
Tumor Markers, Biological/*genetics
Hydroxamic Acids
Tumor Markers, Biological
Azacitidine
Figure
Reference
-
1. Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006. 103:1412–1417.2. Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007. 39:457–466.3. Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007. 3:1709–1723.4. Wood LD, Parsons DW, Jones S, Lin J, Sjóblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007. 318:1108–1113.5. Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003. 3:253–266.6. Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006. 12:989–995.7. deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, Steiger KV, Grützmann R, Pilarsky C, Habermann JK, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009. 55:1337–1346.8. Tänzer M, Balluff B, Distler J, Hale K, Leodolter A, Röcken C, Molnar B, Schmid R, Lofton-Day C, Schuster T, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010. 5:e9061.9. Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, Crispin A, Neumann J, Lamerz R, Kolligs FT. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011. 106:1110–1118.10. Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001. 2:533–543.11. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007. 132:2557–2576.12. Chang MH, You SL, Chen CJ, Liu CJ, Lee CM, Lin SM, Chu HC, Wu TC, Yang SS, Kuo HS, et al. Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: a 20-year follow-up study. J Natl Cancer Inst. 2009. 101:1348–1355.13. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009. 59:225–249.14. Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006. 21:15–21.15. Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007. 27:55–76.16. Kanai Y, Ushijima S, Hui AM, Ochiai A, Tsuda H, Sakamoto M, Hirohashi S. The E-cadherin gene is silenced by CpG methylation in human hepatocellular carcinomas. Int J Cancer. 1997. 71:355–359.17. Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007. 117:2713–2722.18. Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008. 47:908–918.19. Shin SH, Kim BH, Jang JJ, Suh KS, Kang GH. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci. 2010. 25:1152–1159.20. Yuan Y, Wang J, Li J, Wang L, Li M, Yang Z, Zhang C, Dai JL. Frequent epigenetic inactivation of spleen tyrosine kinase gene in human hepatocellular carcinoma. Clin Cancer Res. 2006. 12:6687–6695.21. Lu B, Ma Y, Wu G, Tong X, Guo H, Liang A, Cong W, Liu C, Wang H, Wu M, et al. Methylation of Tip30 promoter is associated with poor prognosis in human hepatocellular carcinoma. Clin Cancer Res. 2008. 14:7405–7412.22. Lee HS, Kim BH, Cho NY, Yoo EJ, Choi M, Shin SH, Jang JJ, Suh KS, Kim YS, Kang GH. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009. 15:812–820.23. Zhang W, Zhou L, Ding SM, Xie HY, Xu X, Wu J, Chen QX, Zhang F, Wei BJ, Eldin AT, et al. Aberrant methylation of the CADM1 promoter is associated with poor prognosis in hepatocellular carcinoma treated with liver transplantation. Oncol Rep. 2011. 25:1053–1062.24. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996. 93:9821–9826.25. Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002. 31:141–149.26. Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002. 21:5483–5495.27. Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006. 125:301–313.28. Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009. 41:178–186.29. Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007. 39:232–236.30. Estécio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, Oki Y, Kondo Y, Jelinek J, Shen L, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. 2010. 20:1369–1382.31. Hereford LM, Osley MA, Ludwig TR 2nd, McLaughlin CS. Cell-cycle regulation of yeast histone mRNA. Cell. 1981. 24:367–375.32. Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002. 14:692–699.33. Nishida H, Suzuki T, Tomaru Y, Hayashizaki Y. A novel replication-independent histone H2a gene in mouse. BMC Genet. 2005. 6:10.34. Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997. 101:284–294.35. Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997. 40:314–322.36. Nishida H, Suzuki T, Ookawa H, Tomaru Y, Hayashizaki Y. Comparative analysis of expression of histone H2a genes in mouse. BMC Genomics. 2005. 6:108.37. Chu M, Tsuda T. Fibulins in development and heritable disease. Birth Defects Res C Embryo Today. 2004. 72:25–36.38. de Vega S, Iwamoto T, Yamada Y. Fibulins: multiple roles in matrix structures and tissue functions. Cell Mol Life Sci. 2009. 66:1890–1902.39. Hucthagowder V, Sausgruber N, Kim KH, Angle B, Marmorstein LY, Urban Z. Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am J Hum Genet. 2006. 78:1075–1080.40. Furuta J, Nobeyama Y, Umebayashi Y, Otsuka F, Kikuchi K, Ushijima T. Silencing of Peroxiredoxin 2 and aberrant methylation of 33 CpG islands in putative promoter regions in human malignant melanomas. Cancer Res. 2006. 66:6080–6086.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Identification of Novel Methylation Markers in Hepatocellular Carcinoma using a Methylation Array
- Detection of Aberrant p16INK4A Methylation in Sera of Patients with Liver Cirrhosis and Hepatocellular Carcinoma
- DNA Methylation in Development
- Molecular Diagnosis and Tumor Markers in Pancreatic Cancer
- A Two-DNA Methylation Signature to Improve Prognosis Prediction of Clear Cell Renal Cell Carcinoma