Allergy Asthma Immunol Res.
2020 Jan;12(1):125-136. 10.4168/aair.2020.12.1.125.
Circulating MicroRNAs and T-Cell Cytokine Expression Are Associated With the Characteristics of Asthma Exacerbation
- Affiliations
-
- 1Department of Immunology and Allergy, Medical University of Lodz, Lodz, Poland. marek.kowalski@csk.umed.lodz.pl
- 2Department of Rheumatology, Medical University of Lodz, Lodz, Poland.
- KMID: 2462577
- DOI: http://doi.org/10.4168/aair.2020.12.1.125
Abstract
- PURPOSE
Immunological mechanisms underlying asthma exacerbation have not been elucidated. The aim of this study was to assess the associations of various asthma exacerbation traits with selected serum microRNA (miRNA) expression and T-cell subpopulations.
METHODS
Twenty-one asthmatics were studied during asthma exacerbation (exacerbation visit [EV] and the follow-up visit [FV] at 6 weeks). At both visits, spirometry was performed, fractional exhaled nitric oxide (FeNO) was measured, and nasopharyngeal and blood samples were collected. In nasopharyngeal samples, respiratory viruses were assayed by multiplex polymerase chain reaction (PCR), and bacterial cultures were performed. Serum miRNAs were assayed with real-time PCR. T-cell surface markers, eosinophil progenitors and intracellular cytokines were assessed by flow cytometry.
RESULTS
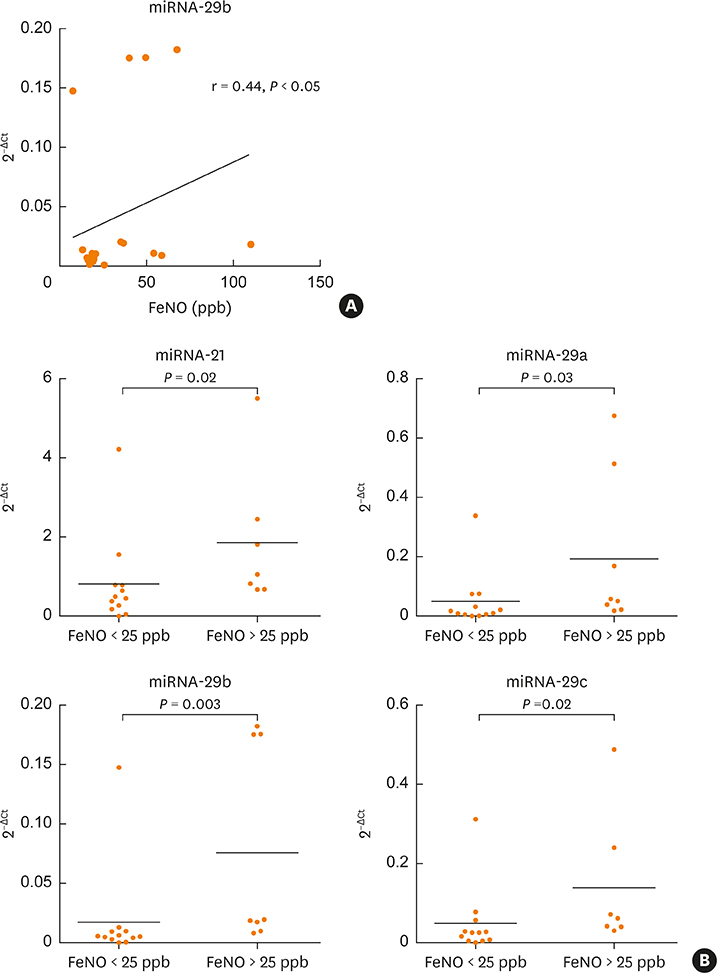
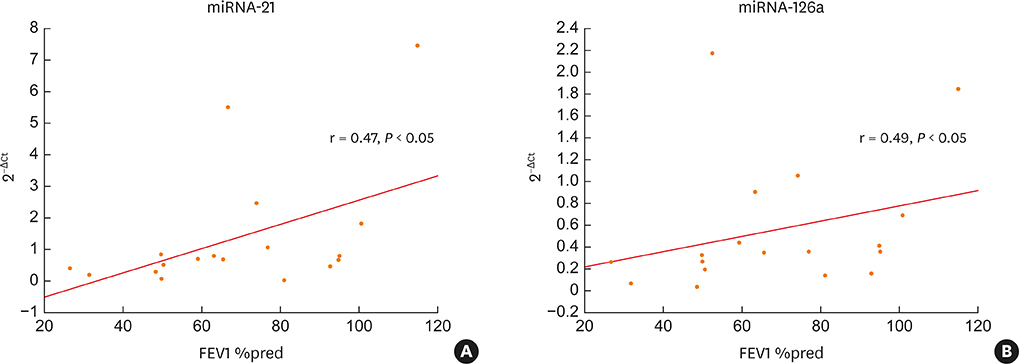
Two-thirds of patients had moderate or severe exacerbation and the FV, overall improvement in asthma control was observed. The mean expression of serum miRNA-126a, miRNA-16 and miRNA-21 was significantly lower at the EV than at the FV. At EV, miRNA-29b correlated with FeNO (r = 0.44, P < 0.05), and 5 of 7 miRNA tested correlated with pulmonary function tests. The number of cluster of differentiation (CD)45+CD4+interleukin (IL)4+ cells was significantly higher at the EV than at the FV, and positive correlations of T-regulatory cells and eosinophil progenitors with asthma control was found. At the EV, serum miRNAs negatively correlated with the number of T cells expressing IL-4, IL-17, IL-22 and interferon gamma, while at the FV both positive and negative correlations with T-cell subsets were observed. No association of detected pathogen (viruses and bacteria) in nasopharyngeal fluid with clinical, functional and immunological parameters was found.
CONCLUSIONS
Epigenetic dysregulation during asthma exacerbation could be related to respiratory function, airway inflammation and T-cell cytokine expression.
Keyword
MeSH Terms
-
Asthma*
Cytokines
Disease Progression
Eosinophils
Epigenomics
Flow Cytometry
Follow-Up Studies
Humans
Inflammation
Interferons
Interleukin-17
Interleukin-4
MicroRNAs*
Multiplex Polymerase Chain Reaction
Nitric Oxide
Real-Time Polymerase Chain Reaction
Respiratory Function Tests
Spirometry
T-Lymphocyte Subsets
T-Lymphocytes*
Cytokines
Interferons
Interleukin-17
Interleukin-4
MicroRNAs
Nitric Oxide
Figure
Reference
-
1. Custovic A, Johnston SL, Pavord I, Gaga M, Fabbri L, Bel EH, et al. EAACI position statement on asthma exacerbations and severe asthma. Allergy. 2013; 68:1520–1531.
Article2. Jackson DJ, Sykes A, Mallia P, Johnston SL. Asthma exacerbations: origin, effect, and prevention. J Allergy Clin Immunol. 2011; 128:1165–1174.
Article3. Farne HA, Johnston SL. Immune mechanisms of respiratory viral infections in asthma. Curr Opin Immunol. 2017; 48:31–37.
Article4. Ghebre MA, Pang PH, Diver S, Desai D, Bafadhel M, Haldar K, et al. Biological exacerbation clusters demonstrate asthma and chronic obstructive pulmonary disease overlap with distinct mediator and microbiome profiles. J Allergy Clin Immunol. 2018; 141:2027–2036.e12.
Article5. Lloyd CM, Hessel EM. Functions of T cells in asthma: more than just T(H)2 cells. Nat Rev Immunol. 2010; 10:838–848.
Article6. Heffler E, Allegra A, Pioggia G, Picardi G, Musolino C, Gangemi S. MicroRNA profiling in asthma: potential biomarkers and therapeutic targets. Am J Respir Cell Mol Biol. 2017; 57:642–650.
Article7. Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016; 164:1226–1232.
Article8. Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012; 129:95–103.
Article9. Levänen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, Pollack JL, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013; 131:894–903.
Article10. Panganiban RP, Pinkerton MH, Maru SY, Jefferson SJ, Roff AN, Ishmael FT. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol. 2012; 1:154–165.11. Maes T, Cobos FA, Schleich F, Sorbello V, Henket M, De Preter K, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol. 2016; 137:1433–1446.
Article12. Kho AT, McGeachie MJ, Moore KG, Sylvia JM, Weiss ST, Tantisira KG. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir Res. 2018; 19:128.
Article13. Stolzenburg LR, Harris A. The role of microRNAs in chronic respiratory disease: recent insights. Biol Chem. 2018; 399:219–234.
Article14. Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009; 182:4994–5002.
Article15. Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A. 2009; 106:18704–18709.
Article16. Lee HY, Lee HY, Choi JY, Hur J, Kim IK, Kim YK, et al. Inhibition of microRNA-21 by an antagomir ameliorates allergic inflammation in a mouse model of asthma. Exp Lung Res. 2017; 43:109–119.
Article17. Liu X, Nelson A, Wang X, Kanaji N, Kim M, Sato T, et al. MicroRNA-146a modulates human bronchial epithelial cell survival in response to the cytokine-induced apoptosis. Biochem Biophys Res Commun. 2009; 380:177–182.
Article18. Global Initiative for Asthma (GINA). Global strategy for asthma management and prevention [Internet]. place unknown: Global Initiative for Asthma;2014. cited 2019 Apr 29. Available from: http://www.ginasthma.org.19. Reddel HK, Taylor DR, Bateman ED, Boulet LP, Boushey HA, Busse WW, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med. 2009; 180:59–99.20. Głobińska A, Pawełczyk M, Kowalski ML. MicroRNAs and the immune response to respiratory virus infections. Expert Rev Clin Immunol. 2014; 10:963–971.
Article21. Perry MM, Adcock IM, Chung KF. Role of microRNAs in allergic asthma: present and future. Curr Opin Allergy Clin Immunol. 2015; 15:156–162.22. Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011; 187:3362–3373.23. Wu XB, Wang MY, Zhu HY, Tang SQ, You YD, Xie YQ. Overexpression of microRNA-21 and microRNA-126 in the patients of bronchial asthma. Int J Clin Exp Med. 2014; 7:1307–1312.24. Hammad Mahmoud Hammad R, Hamed DH, Eldosoky MA, Ahmad AA, Osman HM, Abd Elgalil HM, et al. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 2018; 24:171–179.
Article25. Elbehidy RM, Youssef DM, El-Shal AS, Shalaby SM, Sherbiny HS, Sherief LM, et al. MicroRNA-21 as a novel biomarker in diagnosis and response to therapy in asthmatic children. Mol Immunol. 2016; 71:107–114.
Article26. Wiwanitkit S, Wiwanitkit V. MicroRNA from tuberculosis RNA: a bioinformatics study. J Thorac Dis. 2012; 4:296–297.27. Fang J, Hao Q, Liu L, Li Y, Wu J, Huo X, et al. Epigenetic changes mediated by microRNA miR29 activate cyclooxygenase 2 and lambda-1 interferon production during viral infection. J Virol. 2012; 86:1010–1020.
Article28. Zhang X, Zhao X, Sun H, Yan Y, Huang L, Gu W, et al. The role of miR-29c/B7-H3 axis in children with allergic asthma. J Transl Med. 2018; 16:218.
Article29. Bjornsdottir US, Holgate ST, Reddy PS, Hill AA, McKee CM, Csimma CI, et al. Pathways activated during human asthma exacerbation as revealed by gene expression patterns in blood. PLoS One. 2011; 6:e21902.
Article30. Hansel TT, Tunstall T, Trujillo-Torralbo MB, Shamji B, Del-Rosario A, Dhariwal J, et al. A comprehensive evaluation of nasal and bronchial cytokines and chemokines following experimental rhinovirus infection in allergic asthma: increased interferons (IFN-γ and IFN-λ) and type 2 inflammation (IL-5 and IL-13). EBioMedicine. 2017; 19:128–138.
Article31. Gour N, Wills-Karp M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 2015; 75:68–78.
Article32. Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013; 368:2455–2466.
Article33. Hirose K, Ito T, Nakajima H. Roles of IL-22 in allergic airway inflammation in mice and humans. Int Immunol. 2018; 30:413–418.
Article34. Deliu M, Yavuz TS, Sperrin M, Belgrave D, Sahiner UM, Sackesen C, et al. Features of asthma which provide meaningful insights for understanding the disease heterogeneity. Clin Exp Allergy. 2018; 48:39–47.
Article35. Kowalski ML. Heterogeneity of NSAID-Exacerbated Respiratory Disease: has the time come for subphenotyping? Curr Opin Pulm Med. 2019; 25:64–70.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MicroRNAs in Asthma and Respiratory Infections: Identifying Common Pathways
- Effect of air pollution on acute exacerbation of adult asthma in Seoul, Korea
- The Effects of Bronchiectasis on Asthma Exacerbation
- Seasonality of asthma exacerbation in children caused by respiratory virus infection and allergen sensitization
- Circulating Cell-free Tumor Nucleic Acids in Gastric Cancer