Cancer Res Treat.
2018 Jul;50(3):992-1008. 10.4143/crt.2017.226.
Long Noncoding RNA HEIH Promotes Colorectal Cancer Tumorigenesis via Counteracting miR-939-Mediated Transcriptional Repression of Bcl-xL
- Affiliations
-
- 1Department of General Surgery, Zhujiang Hospital, Southern Medical University, Guangzhou, China. jinlonggyu@163.com
- KMID: 2417888
- DOI: http://doi.org/10.4143/crt.2017.226
Abstract
- PURPOSE
Studies have found that long noncoding RNA HEIH (lncRNA-HEIH) is upregulated and facilitates hepatocellular carcinoma tumor growth. However, its clinical significances, roles, and action mechanism in colorectal cancer (CRC) remains unidentified.
MATERIALS AND METHODS
lncRNA-HEIH expression in CRC tissues and cell lines was measured by quantitative real-time polymerase chain reaction. Cell CountingKit-8, ethynyl deoxyuridine incorporation assay, terminal deoxynucleotidyl transferase dUTP nick end labeling staining, and nude mice xenografts assays were performed to investigate the roles of lncRNA-HEIH. RNA pull-down, RNA immunoprecipitation, chromatin immunoprecipitation, and luciferase reporter assays were performed to investigate the action mechanisms of lncRNA-HEIH.
RESULTS
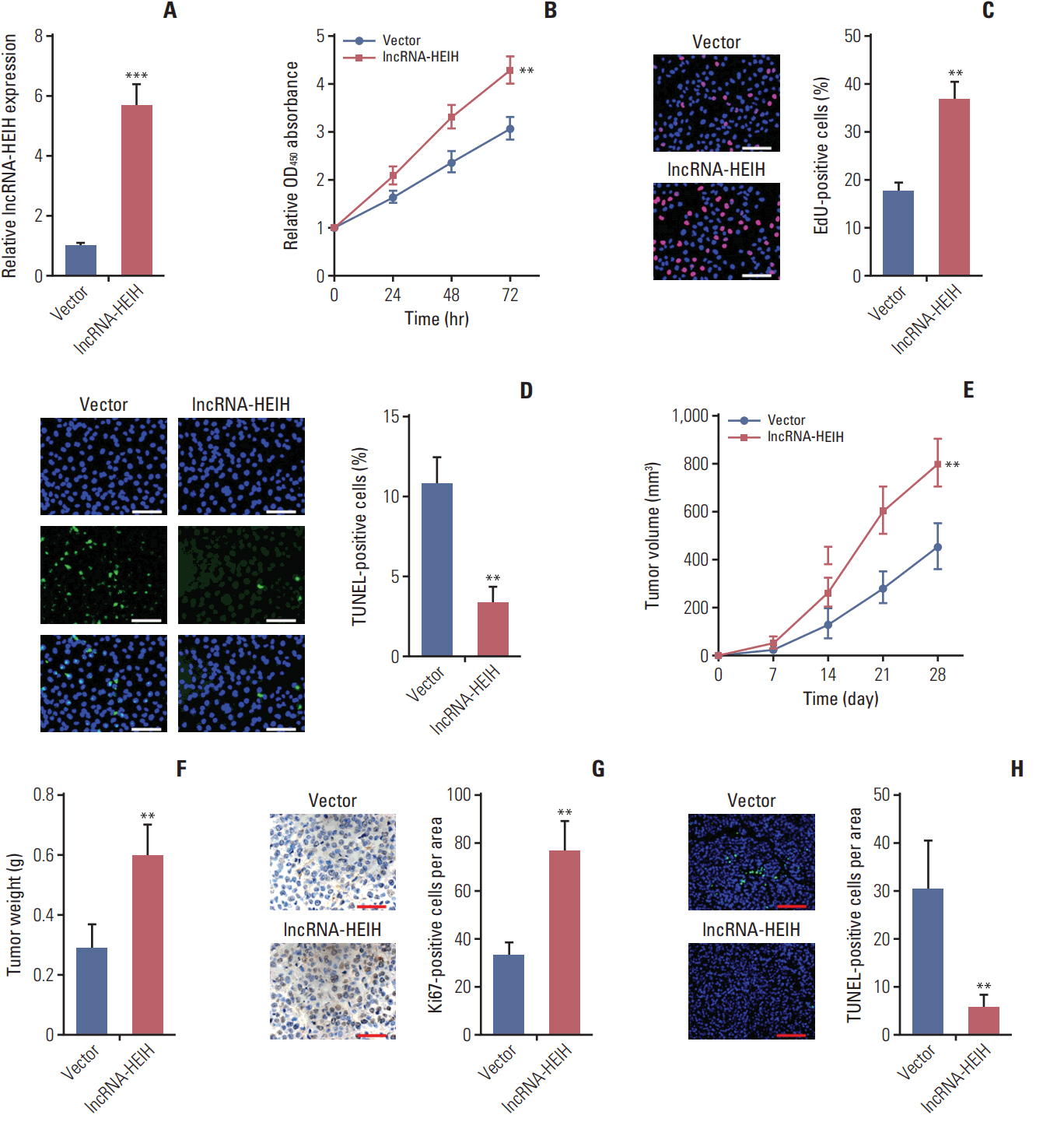
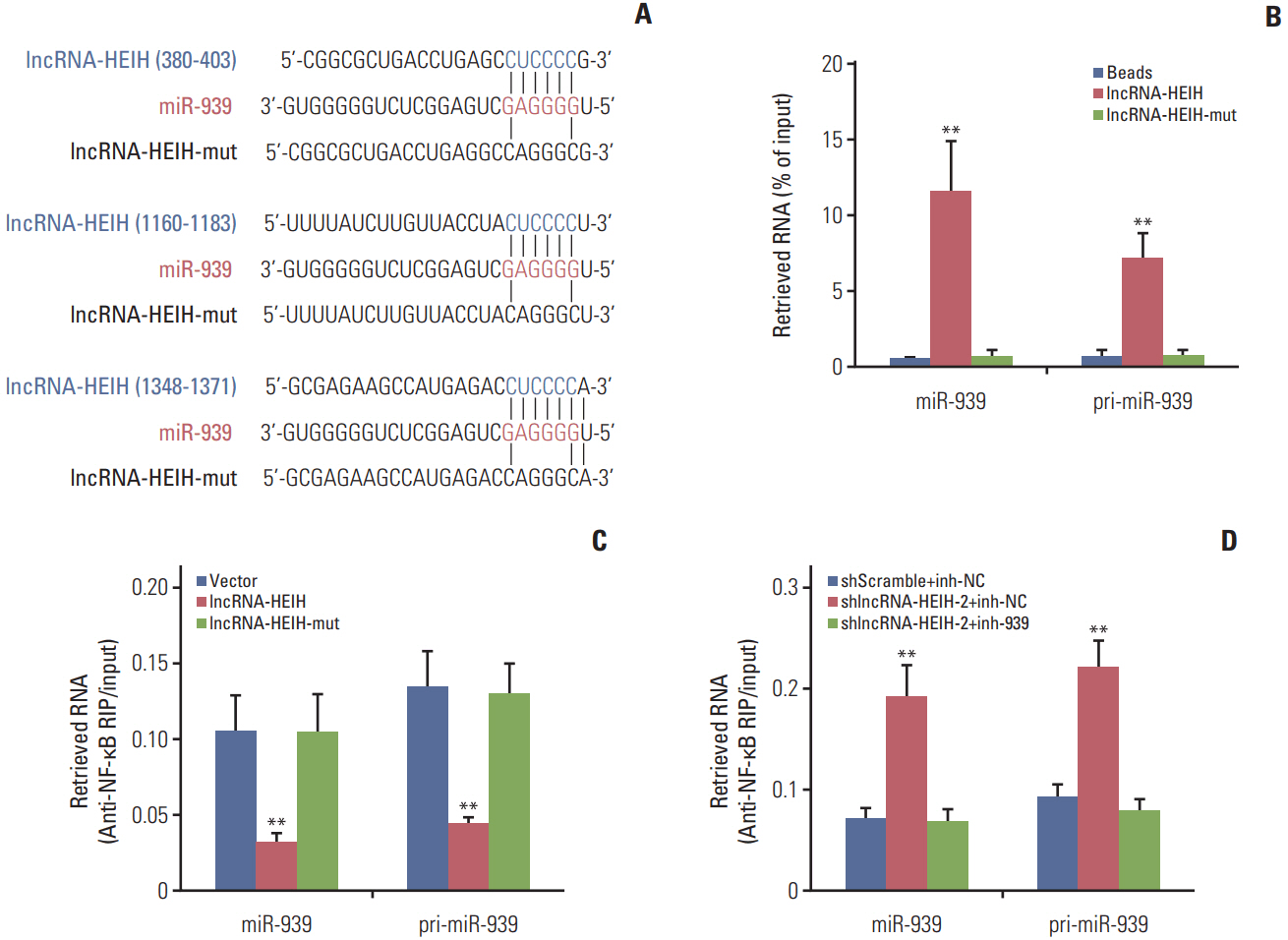
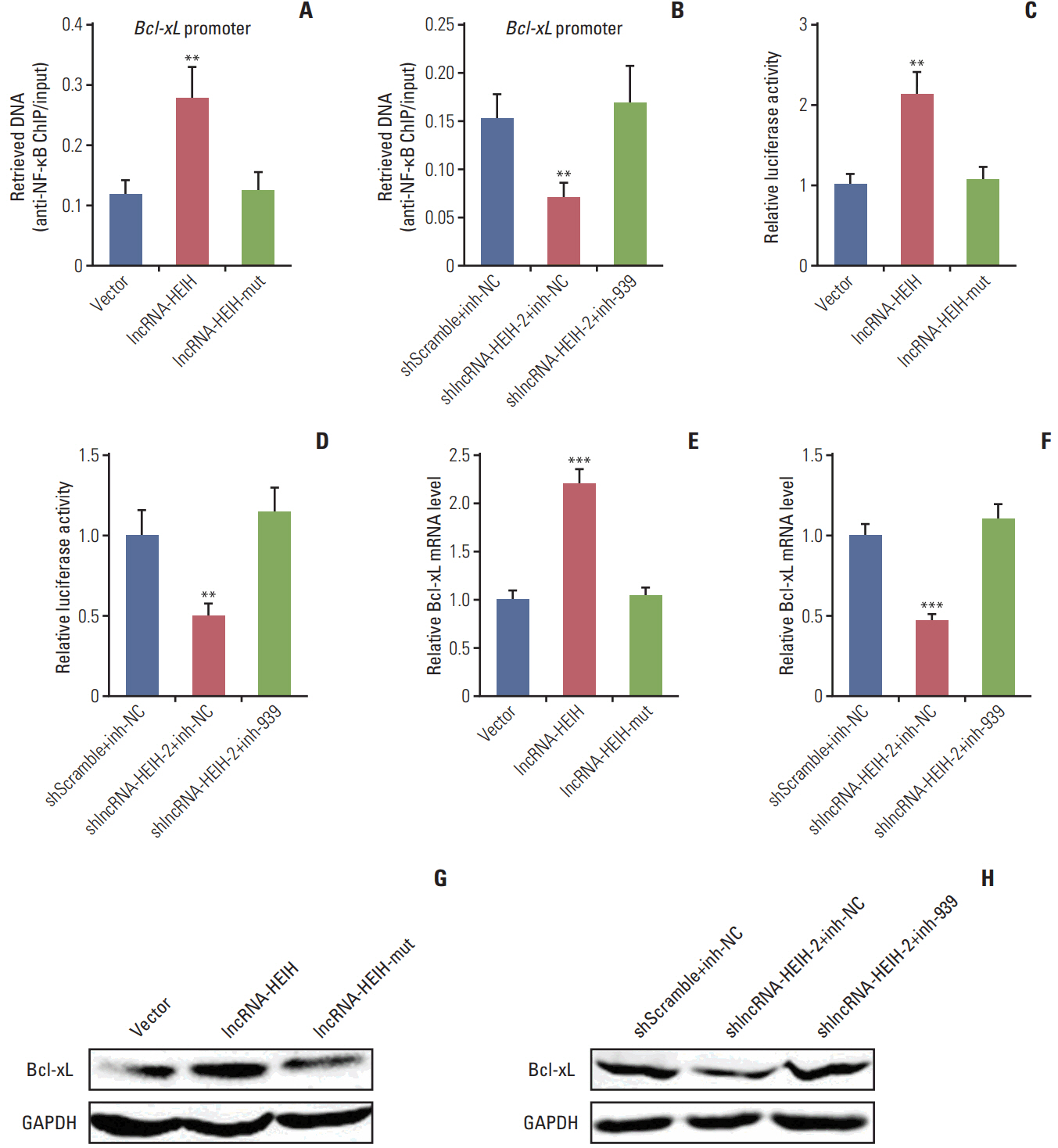
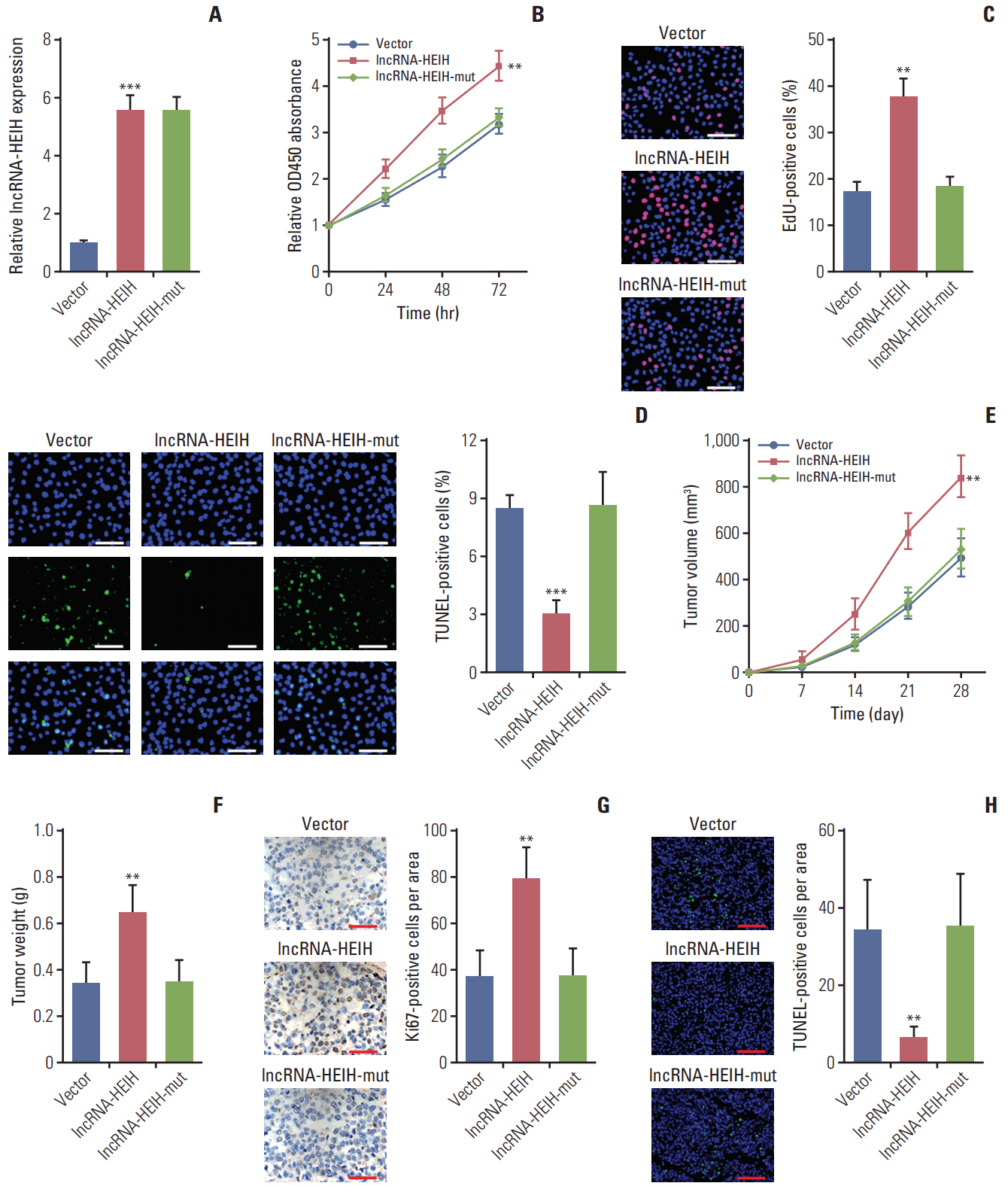
In this study, we found that lncRNA-HEIH is significantly increased in CRC tissues and cell lines. lncRNA-HEIH expression is positively associated with tumor size, invasion depth, and poor prognosis of CRC patients. Enhanced expression of lncRNA-HEIH promotes CRC cell proliferation and decreases apoptosis in vitro, and promotes CRC tumor growth in vivo. Whereas knockdown of lncRNA-HEIH inhibits CRC cell proliferation and induces apoptosis in vitro, and suppresses CRC tumor growth in vivo. Mechanistically, lncRNA-HEIH physically binds to miR-939. The interaction between lncRNA-HEIH and miR-939 damages the binding between miR-939 and nuclear factor κB (NF-κB), increases the binding of NF-κB to Bcl-xL promoter, and promotes the transcription and expression of Bcl-xL. Moreover, Bcl-xL expression is positively associatedwith lncRNA-HEIH in CRC tissues. Blocking the interaction between lncRNA-HEIH and miR-939 abolishes the effects of lncRNA-HEIH on CRC tumorigenesis.
CONCLUSION
This study demonstrated that lncRNA-HEIH promotes CRC tumorigenesis through counteracting miR-939-mediated transcriptional repression of Bcl-xL, and suggested that lncRNA-HEIH may serve as a prognostic biomarker and therapeutic target for CRC.
Keyword
MeSH Terms
-
Animals
Apoptosis
Carcinogenesis*
Carcinoma, Hepatocellular
Cell Line
Cell Proliferation
Chromatin Immunoprecipitation
Colorectal Neoplasms*
Deoxyuridine
DNA Nucleotidylexotransferase
Heterografts
Humans
Immunoprecipitation
In Vitro Techniques
Luciferases
Mice
Mice, Nude
Prognosis
Real-Time Polymerase Chain Reaction
Repression, Psychology*
RNA
RNA, Long Noncoding*
DNA Nucleotidylexotransferase
Deoxyuridine
Luciferases
RNA
RNA, Long Noncoding
Figure
Reference
-
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.
Article2. He D, Ma L, Feng R, Zhang L, Jiang Y, Zhang Y, et al. Analyzing large-scale samples highlights significant association between rs10411210 polymorphism and colorectal cancer. Biomed Pharmacother. 2015; 74:164–8.
Article3. Levy J, Cacheux W, Bara MA, L'Hermitte A, Lepage P, Fraudeau M, et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015; 17:1062–73.
Article4. Li Y, Liang L, Dai W, Cai G, Xu Y, Li X, et al. Prognostic impact of programed cell death-1 (PD-1) and PD-ligand 1 (PD-L1) expression in cancer cells and tumor infiltrating lymphocytes in colorectal cancer. Mol Cancer. 2016; 15:55.
Article5. Depeille P, Henricks LM, van de Ven RA, Lemmens E, Wang CY, Matli M, et al. RasGRP1 opposes proliferative EGFRSOS1-Ras signals and restricts intestinal epithelial cell growth. Nat Cell Biol. 2015; 17:804–15.
Article6. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009; 361:2449–60.7. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012; 489:101–8.
Article8. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasionmetastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014; 25:666–81.9. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009; 10:155–9.
Article10. Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016; 18:431–42.
Article11. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triplenegative breast cancer. Nat Cell Biol. 2016; 18:213–24.12. Rui Q, Xu Z, Yang P, He Z. Long noncoding RNA expression patterns in lymph node metastasis in colorectal cancer by microarray. Biomed Pharmacother. 2015; 75:12–8.
Article13. Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011; 54:1679–89.
Article14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–97.15. Ge Y, Zhang L, Nikolova M, Reva B, Fuchs E. Strand-specific in vivo screen of cancer-associated miRNAs unveils a role for miR-21(*) in SCC progression. Nat Cell Biol. 2016; 18:111–21.16. Dror S, Sander L, Schwartz H, Sheinboim D, Barzilai A, Dishon Y, et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat Cell Biol. 2016; 18:1006–17.
Article17. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011; 54:2025–35.
Article18. Oliverio M, Schmidt E, Mauer J, Baitzel C, Hansmeier N, Khani S, et al. Dicer1-miR-328-Bace1 signalling controls brown adipose tissue differentiation and function. Nat Cell Biol. 2016; 18:328–36.
Article19. Cui C, Yu J, Huang S, Zhu H, Huang Z. Transcriptional regulation of gene expression by microRNAs as endogenous decoys of transcription factors. Cell Physiol Biochem. 2014; 33:1698–714.
Article20. Chen HC, Kanai M, Inoue-Yamauchi A, Tu HC, Huang Y, Ren D, et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family. Nat Cell Biol. 2015; 17:1270–81.
Article21. Zheng X, Zhang Y, Zhang L, Xu W, Ma W, Sun R, et al. Synergistic inhibition of sunitinib and ethaselen against human colorectal cancer cells proliferation. Biomed Pharmacother. 2016; 83:212–20.
Article22. Feng M, Feng J, Chen W, Wang W, Wu X, Zhang J, et al. Lipocalin2 suppresses metastasis of colorectal cancer by attenuating NF-kappaB-dependent activation of snail and epithelial mesenchymal transition. Mol Cancer. 2016; 15:77.
Article23. Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016; 80:73–9.
Article24. Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016; 283:3739–54.
Article25. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008; 105:1608–13.
Article26. Zhang JX, Xu Y, Gao Y, Chen C, Zheng ZS, Yun M, et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017; 16:18.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Long Non-Coding RNA LINC00525 Promotes the Stemness and Chemoresistance of Colorectal Cancer by Targeting miR-507/ELK3 Axis
- LncRNA XLOC_006390 facilitates cervical cancer tumorigenesis and metastasis as a ceRNA against miR-331-3p and miR-338-3p
- Long non-coding RNA linc00152 acting as a promising oncogene in cancer progression
- MiR-4492, a New Potential MicroRNA for Cancer Diagnosis and Treatment: A Mini Review
- Long Noncoding-RNA Component of Mitochondrial RNA Processing Endoribonuclease Promotes Carcinogenesis in Triple-Negative Breast Cancer Cells via the Competing Endogenous RNA Mechanism