Int J Stem Cells.
2019 Jul;12(2):347-359. 10.15283/ijsc19041.
Long Non-Coding RNA LINC00525 Promotes the Stemness and Chemoresistance of Colorectal Cancer by Targeting miR-507/ELK3 Axis
- Affiliations
-
- 1Department of Colorectal Surgery, Yidu Central Hospital of Weifang City, Qingzhou, China. wangshunsheng728@126.com
- KMID: 2465904
- DOI: http://doi.org/10.15283/ijsc19041
Abstract
- BACKGROUND AND OBJECTIVES
This study aims to explore the effects of a long non-coding RNA, LINC00525, on colorectal cancer (CRC) and its underlying molecular mechanisms.
METHODS
The qPCR, MTT, colony formation, Western blotting, Luciferase reporter and biotin pull-down, shRNA knockdown and DNA fragmentation assays were performed in this study.
RESULTS
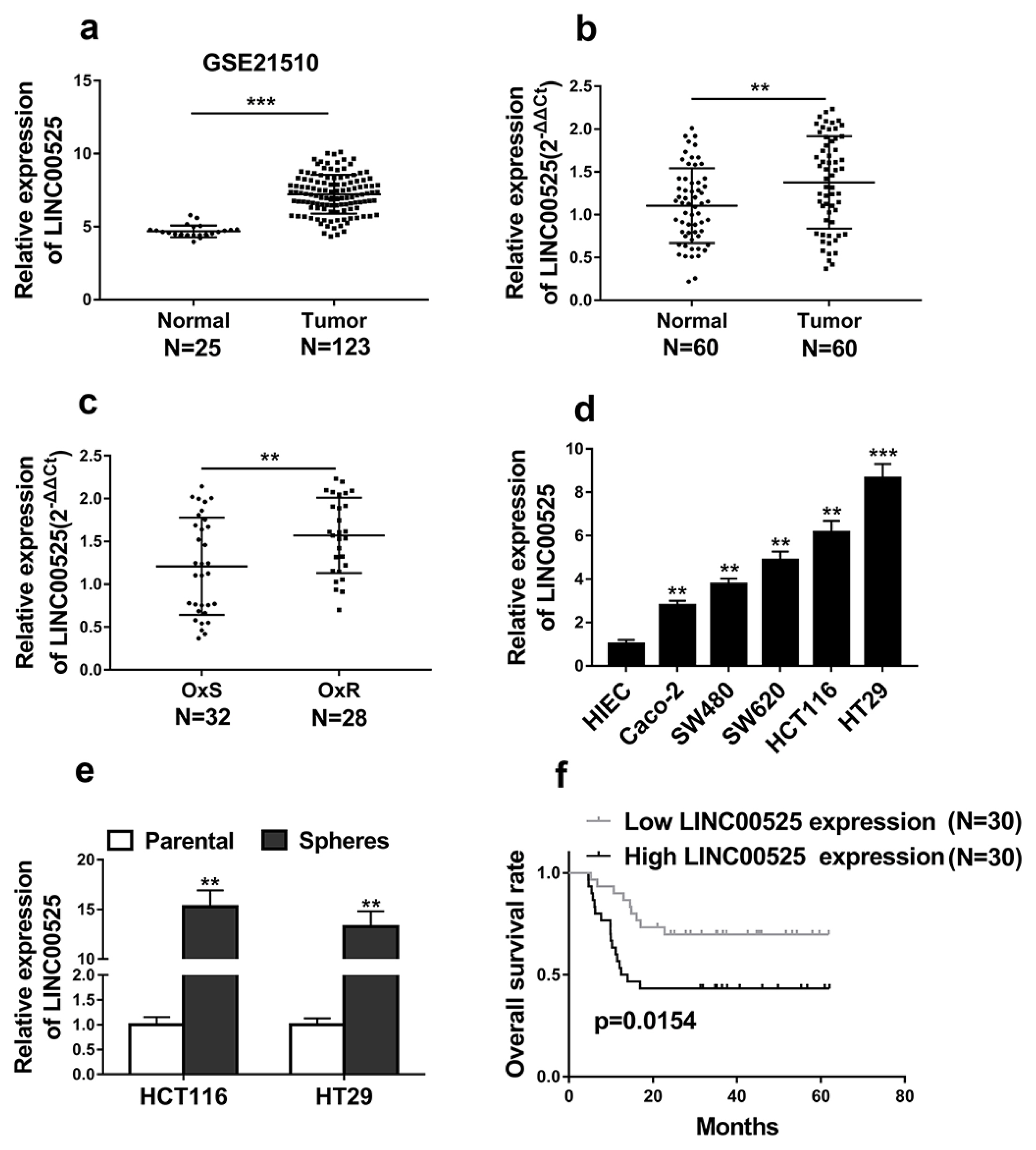
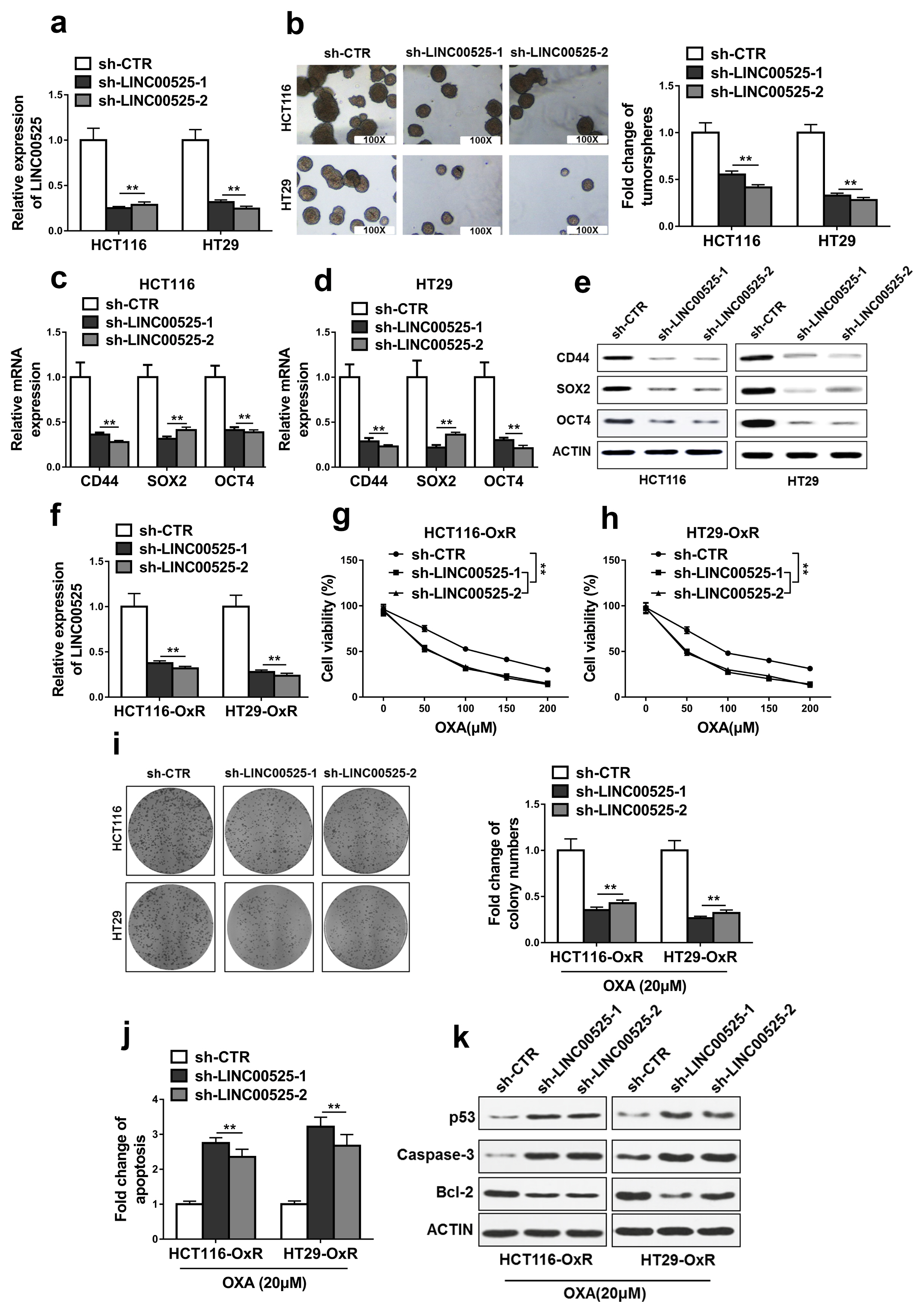
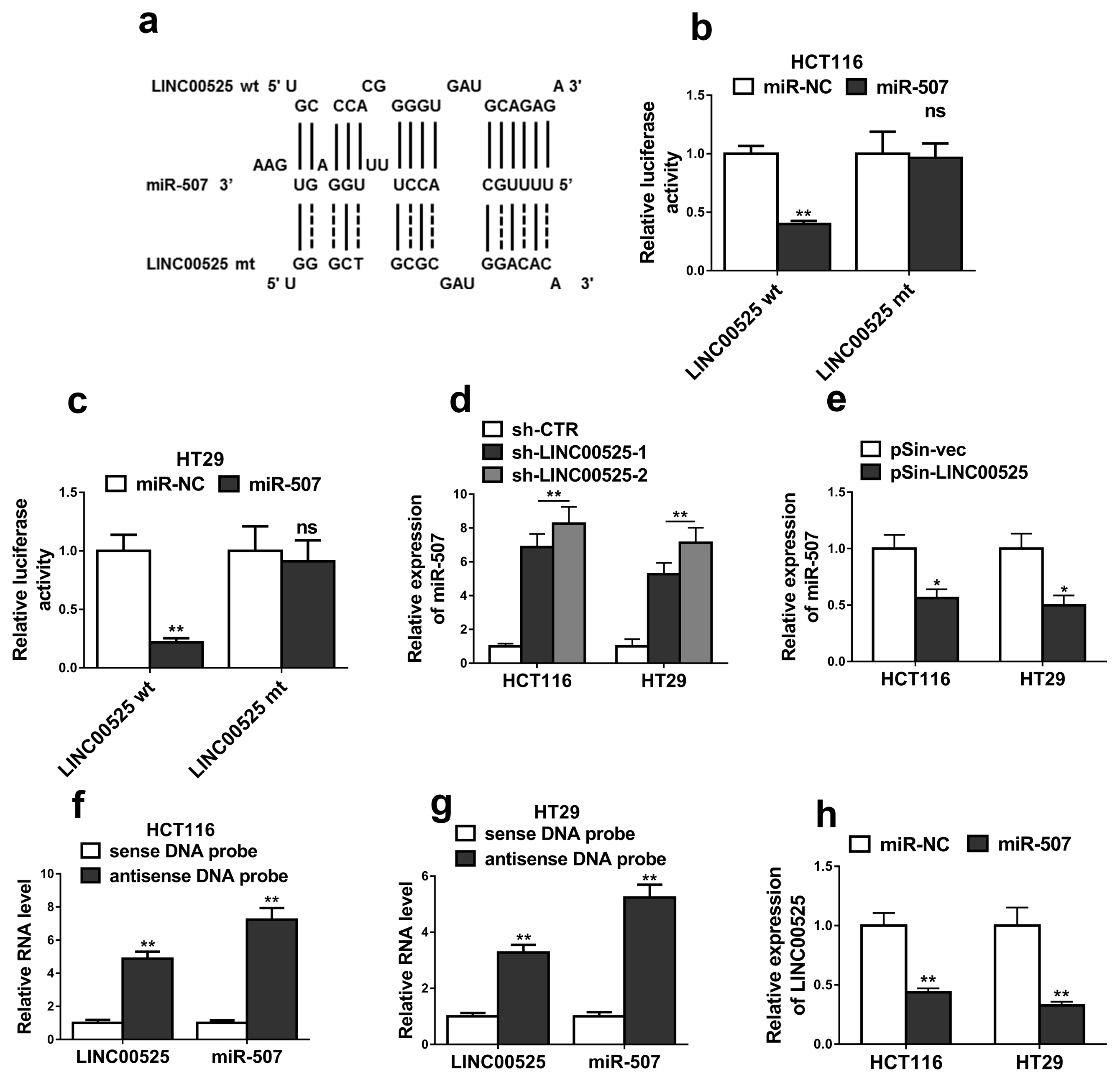
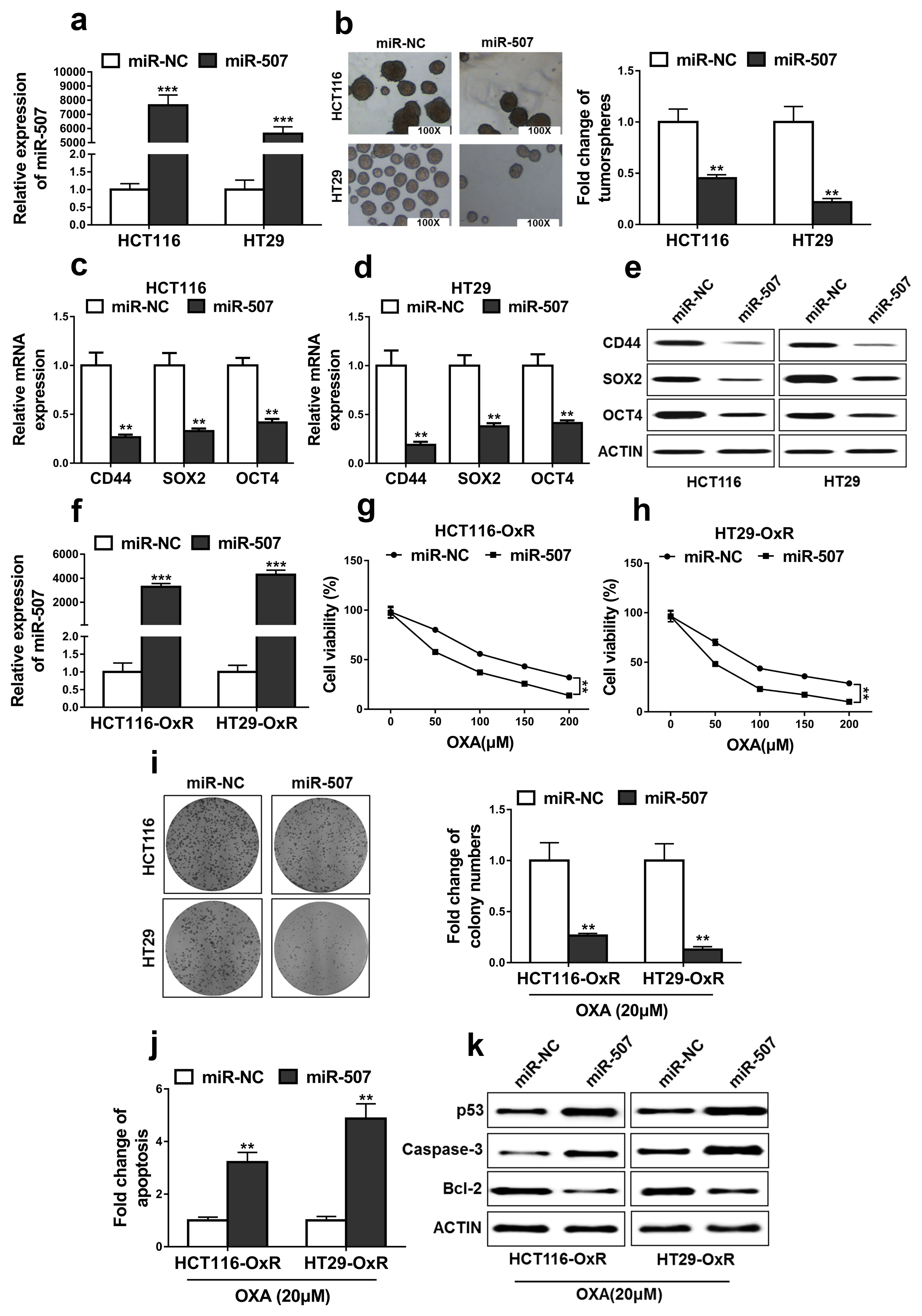
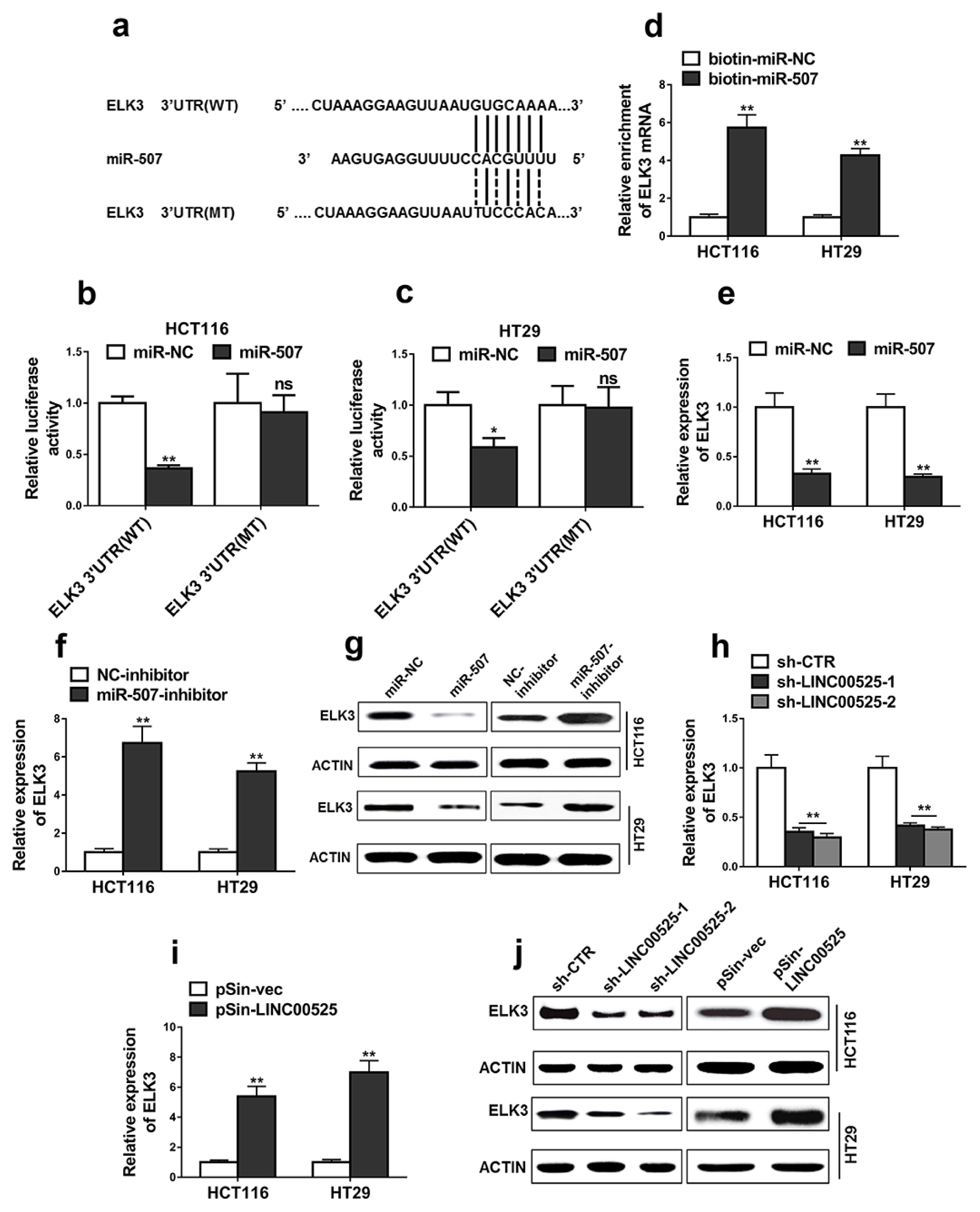
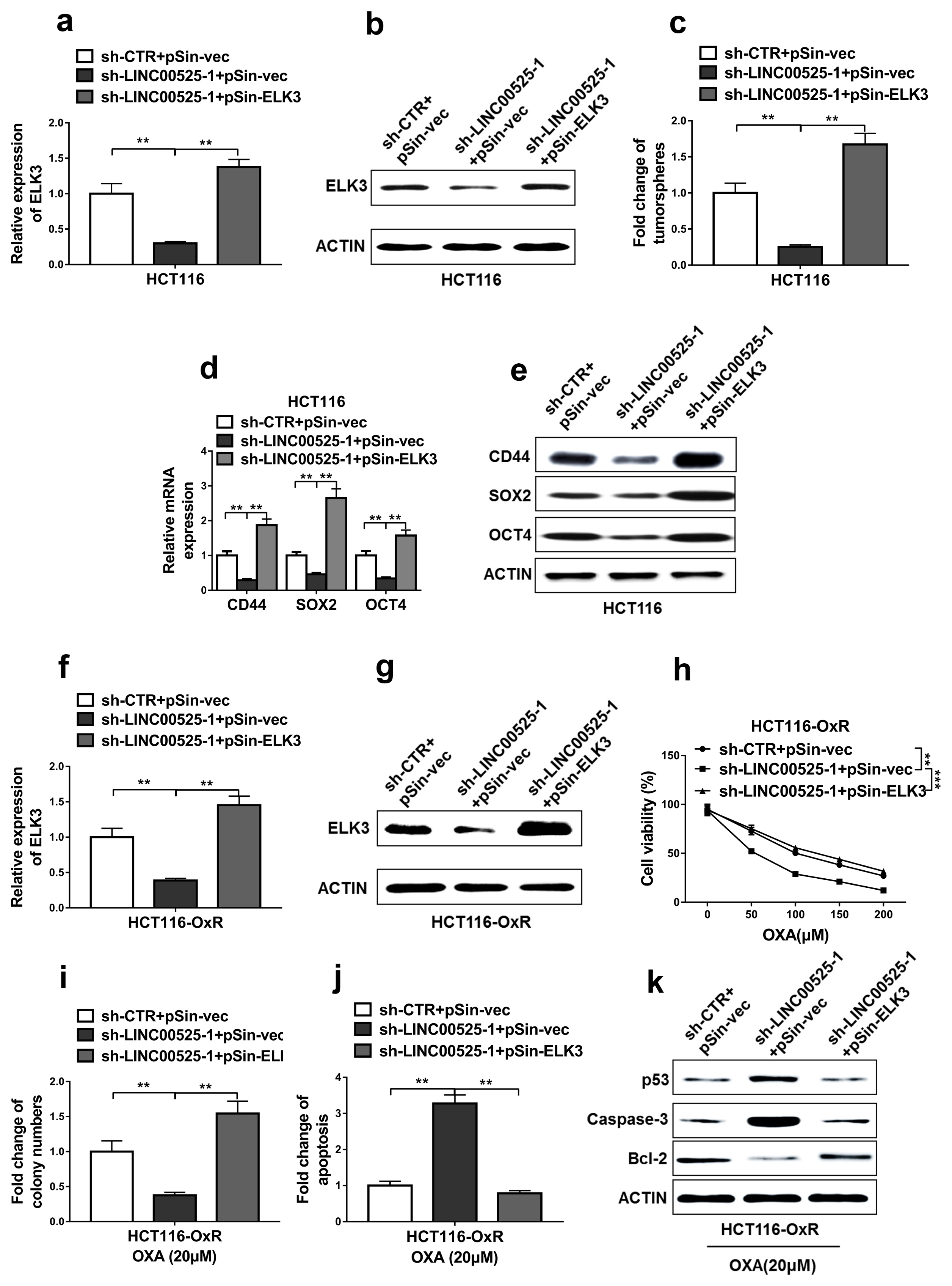
High expressions of LINC00525 were associated with poor prognosis of CRC patients. LINC00525 knockdown decreased stemness properties and increased sensitivities to oxaliplatin. MiR-507 was a direct target of LINC00525 and overexpression of miR-507 significantly decreased abilities of tumorsphere formation and cell growth. Overexpression of miR-507 resulted in a decrease of expression of cancer stem cell markers and the increase of apoptosis rates. MiR-507 regulated the expression of ELK3. In addition, LINC00525 knockdown decreased the expression of ELK3. Restoration of ELK3 expression abrogated the effects of LINC00525 knockdown. LINC00525 could be served as prognostic marker of CRC.
CONCLUSIONS
LINC00525 enhanced stemness properties and increased sensitivities of CRC cells to oxaliplatin by targeting miR-507/ELK3 axis.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Midgley R, Kerr D. Immunotherapy for colorectal cancer: a challenge to clinical trial design. Lancet Oncol. 2000; 1:159–168. DOI: 10.1016/S1470-2045(00)00034-6. PMID: 11905654.
Article2. Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005; 352:476–487. DOI: 10.1056/NEJMra040958. PMID: 15689586.
Article3. Hwang IG, Jang JS, Oh SY, Rho MH, Lee S, Park YS, Park JO, Nam EM, Lee HR, Jun HJ, Chi KC. Phase II study of mFOLFOX3 (5-fluorouracil, leucovorin, oxaliplatin) as second-line treatment after gemcitabine failure in patients with unresectable/metastatic biliary tract cancer. Cancer Chemother Pharmacol. 2015; 75:757–762. DOI: 10.1007/s00280-015-2691-1. PMID: 25677446.
Article4. Chocry M, Leloup L, Kovacic H. Correction: reversion of resistance to oxaliplatin by inhibition of p38 MAPK in colorectal cancer cell lines: involvement of the calpain/Nox1 pathway. Oncotarget. 2018; 9:26978–26979. DOI: 10.18632/oncotarget.25605. PMID: 29928496. PMCID: PMC6003572.
Article5. Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019; 24:41–53. DOI: 10.1016/j.stem.2018.12.009. PMID: 30609398. PMCID: PMC6350931.
Article6. Al-Alem LF, Pandya UM, Baker AT, Bellio C, Zarrella BD, Clark J, DiGloria CM, Rueda BR. Ovarian cancer stem cells: what progress have we made? Int J Biochem Cell Biol. 2019; 107:92–103. DOI: 10.1016/j.biocel.2018.12.010. PMID: 30572025.
Article7. Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005; 5:275–284. DOI: 10.1038/nrc1590. PMID: 15803154.
Article8. Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer Stem Cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018; 2018:5416923. DOI: 10.1155/2018/5416923. PMID: 29681949. PMCID: PMC5850899.
Article9. Hon KW, Abu N, Ab Mutalib NS, Jamal R. miRNAs and lncRNAs as predictive biomarkers of response to FOLFOX therapy in colorectal cancer. Front Pharmacol. 2018; 9:846. DOI: 10.3389/fphar.2018.00846. PMID: 30127741. PMCID: PMC6088237.
Article10. Li CH, Chen Y. Diagnostic potential of lncRNAs in cancer. EBioMedicine. 2016; 7:7–8. DOI: 10.1016/j.ebiom.2016.05.013. PMID: 27322445. PMCID: PMC4909482.
Article11. Chen S, Zhu J, Wang F, Guan Z, Ge Y, Yang X, Cai J. LncRNAs and their role in cancer stem cells. Oncotarget. 2017; 8:110685–110692. DOI: 10.18632/oncotarget.22161. PMID: 29299179. PMCID: PMC5746414.
Article12. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr). 2018; 41:585–603. DOI: 10.1007/s13402-018-0406-4. PMID: 30218296.
Article13. Xu Z, Wang C, Xiang X, Li J, Huang J. Characterization of mRNA expression and endogenous RNA profiles in bladder cancer based on The Cancer Genome Atlas (TCGA) database. Med Sci Monit. 2019; 25:3041–3060. DOI: 10.12659/MSM.915487. PMID: 31020952. PMCID: PMC6498884.
Article14. Wang J, Zhang Y, Wan H, Dong N, Bao L, Sun X, Xu M, Wang X. Global analyses of subtype- or stage-specific genes on chromosome 7 in patients with lung cancer. Cancer Metastasis Rev. 2015; 34:333–345. DOI: 10.1007/s10555-015-9568-y. PMID: 25951983.
Article15. Labaj W, Papiez A, Polanska J, Planski A. Deep data analysis of a large microarray collection for leukemia biomarker identification. Paper presented at: 10th International Conference on Practical Applications of Computational Biology & Bioinformatics. 2016 Jun 1–3; Seville, Spain.
Article16. Liu L, Liu J, Wang H, Zhao H, Du Y. Fenretinide targeting of human colon cancer sphere cells through cell cycle regulation and stress-responsive activities. Oncol Lett. 2018; 16:5339–5348. PMID: 30250604. PMCID: PMC6144862.
Article17. Yang H, Liu C, Zhang YQ, Ge LT, Chen J, Jia XQ, Gu RX, Sun Y, Sun WD. Ilexgenin A induces B16–F10 melanoma cell G1/S arrest in vitro and reduces tumor growth in vivo. Int Immunopharmacol. 2015; 24:423–431. DOI: 10.1016/j.intimp.2014.12.040. PMID: 25596038.
Article18. Xu K, Chen G, Qiu Y, Yuan Z, Li H, Yuan X, Sun J, Xu J, Liang X, Yin P. miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget. 2017; 8:21719–21732. PMID: 28423513. PMCID: PMC5400618.
Article19. Yamamoto K, Ito S, Hanafusa H, Shimizu K, Ouchida M. Uncovering direct targets of MiR-19a involved in lung cancer progression. PLoS One. 2015; 10:e0137887. DOI: 10.1371/journal.pone.0137887. PMID: 26367773. PMCID: PMC4569347.
Article20. Shang W, Jiang Y, Boettcher M, Ding K, Mollenauer M, Liu Z, Wen X, Liu C, Hao P, Zhao S, McManus MT, Wei L, Weiss A, Wang H. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc Natl Acad Sci U S A. 2018; 115:E4051–E4060. DOI: 10.1073/pnas.1801340115. PMID: 29632189. PMCID: PMC5924929.
Article21. Bruera G, Ricevuto E. Intensive chemotherapy of metastatic colorectal cancer: weighing between safety and clinical efficacy: evaluation of Masi G, Loupakis F, Salvatore L, et al. Bevacizumab with FOLFOXIRI (irinotecan, oxaliplatin, fluorouracil, and folinate) as first-line treatment for metastatic colorectal cancer: a phase 2 trial. Lancet Oncol. 2011; 11:821–824. DOI: 10.1517/14712598.2011.582462. PMID: 21545334.
Article22. Becouarn Y, Rougier P. Clinical efficacy of oxaliplatin monotherapy: phase II trials in advanced colorectal cancer. Semin Oncol. 1998; 25(2 Suppl 5):23–31. PMID: 9609105.23. deBraud F, Munzone E, Nolè F, De Pas T, Biffi R, Brienza S, Aapro MS. Synergistic activity of oxaliplatin and 5-fluorouracil in patients with metastatic colorectal cancer with progressive disease while on or after 5-fluorouracil. Am J Clin Oncol. 1998; 21:279–283. DOI: 10.1097/00000421-199806000-00015. PMID: 9626798.
Article24. Comella P, Casaretti R, Sandomenico C, Avallone A, Franco L. Role of oxaliplatin in the treatment of colorectal cancer. Ther Clin Risk Manag. 2009; 5:229–238. DOI: 10.2147/TCRM.S3583. PMID: 19436599. PMCID: PMC2697505.
Article25. Tian L, Hu X, He Y, Wu Z, Li D, Zhang H. Construction of lncRNA-miRNA-mRNA networks reveals functional lncRNAs in abdominal aortic aneurysm. Exp Ther Med. 2018; 16:3978–3986. PMID: 30344676. PMCID: PMC6176170.
Article26. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016; 1402:271–286. DOI: 10.1007/978-1-4939-3378-5_21. PMID: 26721498.
Article27. Guo H, Qi RQ, Sheng J, Liu C, Ma H, Wang HX, Li JH, Gao XH, Wan YS, Chen HD. MiR-155, a potential serum marker of extramammary Paget’s disease. BMC Cancer. 2018; 18:1078. DOI: 10.1186/s12885-018-4994-1. PMID: 30458743. PMCID: PMC6247506.
Article28. Jia L, Liu W, Cao B, Li H, Yin C. MiR-507 inhibits the migration and invasion of human breastcancer cells through Flt-1 suppression. Oncotarget. 2016; 7:36743–36754. DOI: 10.18632/oncotarget.9163. PMID: 27167339. PMCID: PMC5095036.
Article29. Wang LH, Lin CY, Liu SC, Liu GT, Chen YL, Chen JJ, Chan CH, Lin TY, Chen CK, Xu GH, Chen SS, Tang CH, Wang SW. CCL5 promotes VEGF-C production and induces lymphangiogenesis by suppressing miR-507 in human chondrosarcoma cells. Oncotarget. 2016; 7:36896–36908. PMID: 27166194. PMCID: PMC5095047.
Article30. Gross C, Dubois-Pot H, Wasylyk B. The ternary complex factor Net/Elk-3 participates in the transcriptional response to hypoxia and regulates HIF-1 alpha. Oncogene. 2008; 27:1333–1341. DOI: 10.1038/sj.onc.1210736. PMID: 17704799.
Article31. Wasylyk C, Zheng H, Castell C, Debussche L, Multon MC, Wasylyk B. Inhibition of the Ras-Net (Elk-3) pathway by a novel pyrazole that affects microtubules. Cancer Res. 2008; 68:1275–1283. DOI: 10.1158/0008-5472.CAN-07-2674. PMID: 18316589.
Article32. Lee JH, Hur W, Hong SW, Kim JH, Kim SM, Lee EB, Yoon SK. ELK3 promotes the migration and invasion of liver cancer stem cells by targeting HIF-1α. Oncol Rep. 2017; 37:813–822. DOI: 10.3892/or.2016.5293. PMID: 27959451.
Article33. Park JH, Kim KP, Ko JJ, Park KS. PI3K/Akt/mTOR activation by suppression of ELK3 mediates chemosensitivity of MDA-MB-231 cells to doxorubicin by inhibiting autophagy. Biochem Biophys Res Commun. 2016; 477:277–282. DOI: 10.1016/j.bbrc.2016.06.057. PMID: 27301639.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Long Noncoding RNA PVT1 Promotes Stemness and Temozolomide Resistance through miR-365/ELF4/SOX2 Axis in Glioma
- MEG3 LncRNA from Exosomes Released from Cancer-Associated Fibroblasts Enhances Cisplatin Chemoresistance in SCLC via a MiR-15a-5p/CCNE1 Axis
- LncRNA STARD7-AS1 suppresses cervical cancer cell proliferation while promoting autophagy by regulating miR-31-5p/TXNIP axis to inactivate the mTOR signaling
- Long Non-coding RNA CCAT1 Sponges miR-454 to Promote Chemoresistance of Ovarian Cancer Cells to Cisplatin by Regulation of Surviving
- Ribosomal Protein L9 Maintains Stemness of Colorectal Cancer via an ID-1 Dependent Mechanism