J Korean Med Sci.
2016 May;31(5):688-694. 10.3346/jkms.2016.31.5.688.
Silencing of Long Non-Coding RNA MALAT1 Promotes Apoptosis of Glioma Cells
- Affiliations
-
- 1Department of Neurology, Yishui Central Hospital, Shandong, China.
- 2Department of Oncology, Yantai Yuhuangding Hospital Affiliated to Qingdao University, Shandong, China. kiwi_2009@163.com
- KMID: 2373664
- DOI: http://doi.org/10.3346/jkms.2016.31.5.688
Abstract
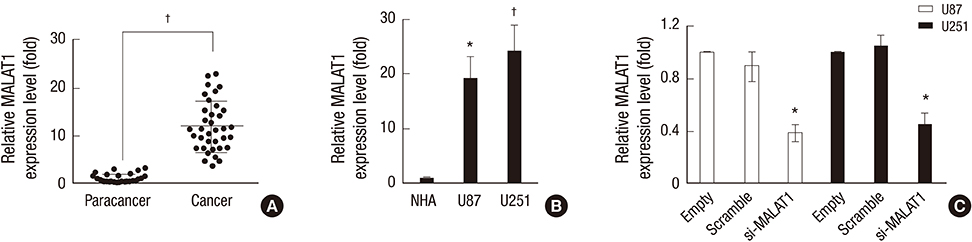
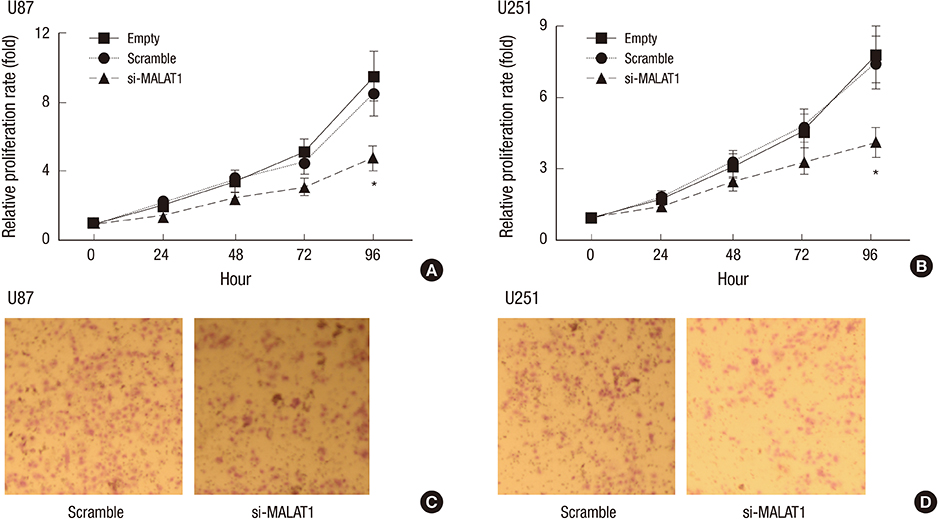
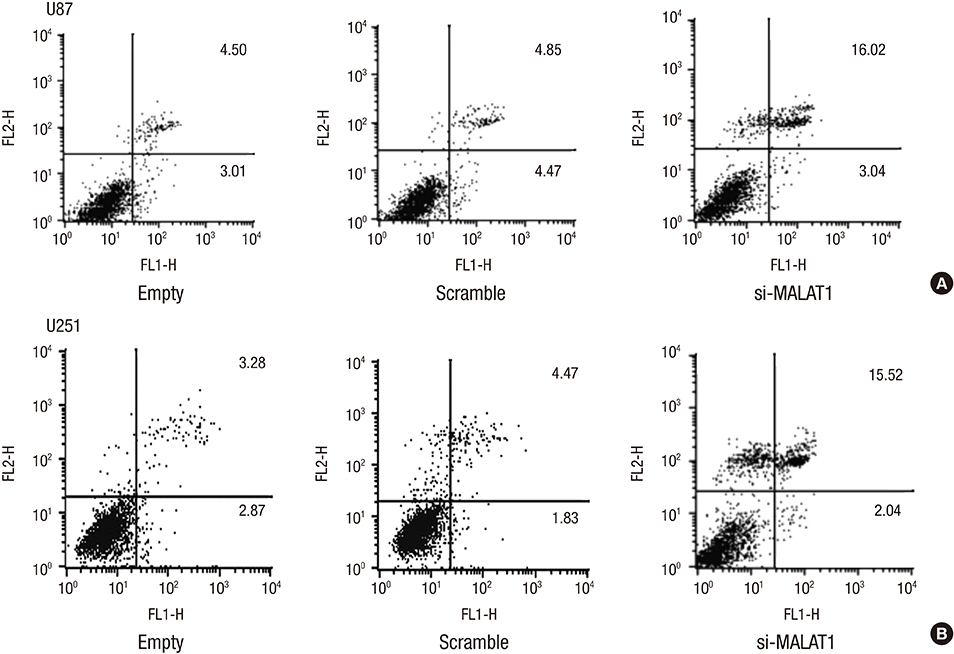
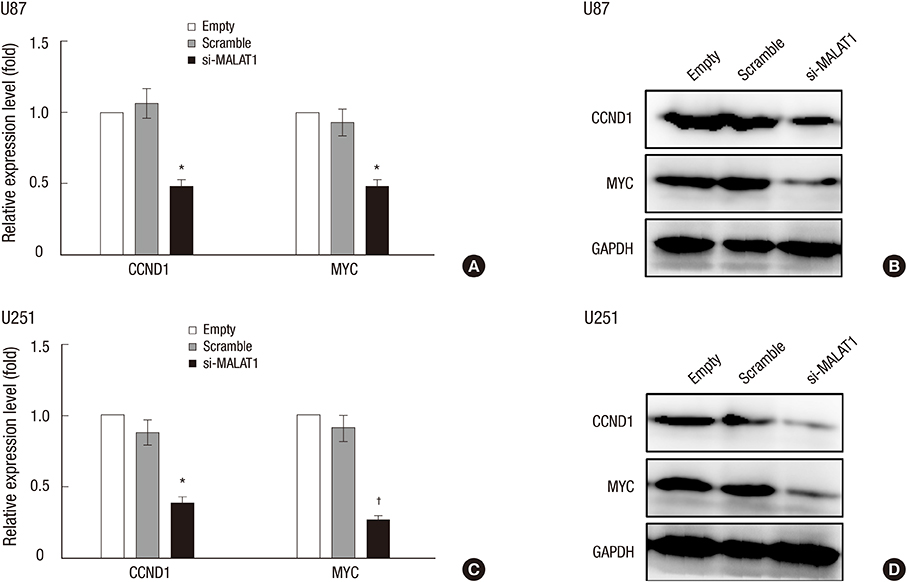
- The metastasis-associated lung adenocarcinoma transcription 1 (MALAT1) is a highly conserved long non-coding RNA (lncRNA) gene. However, little is known about the pathological role of lncRNA MALAT1 in glioma. In the present study, we explored the expression level of lncRNA MALAT1 in primary glioma tissues as well as in U87 and U251 glioma cell lines. Using qRT-PCR, we found that the expression of lncRNA MALAT1 was significantly increased in glioma tissues compared with that of paracancerous tissues. Meanwhile, the expression of MALAT1 was highly expressed in U98 and U251 cells. In order to explore the function of MALAT1, the expression of MALAT1 was greatly reduced in U87 and U251 cells transfected with siRNA specifically targeting MALAT1. Consequently, cell viability of U87 and U251 cells were drastically decreased after the knockdown of MALAT1. Concomitantly, the apoptosis rate of the two cell lines was dramatically increased. Furthermore, the expression levels of some tumor markers were reduced after the knockdown of MALAT1, such as CCND1 and MYC. In summary, the current study indicated a promoting role of MALAT1 in the development of glioma cell.
MeSH Terms
-
*Apoptosis
Biomarkers, Tumor/genetics/metabolism
Blotting, Western
Cell Line, Tumor
Cell Movement
Cell Proliferation
Cyclin D1/genetics/metabolism
Down-Regulation
Flow Cytometry
Glioma/metabolism/pathology
Humans
Proto-Oncogene Proteins c-myc/genetics/metabolism
*RNA Interference
RNA, Long Noncoding/antagonists & inhibitors/genetics/*metabolism
RNA, Small Interfering/metabolism
Real-Time Polymerase Chain Reaction
Biomarkers, Tumor
Cyclin D1
Proto-Oncogene Proteins c-myc
RNA, Long Noncoding
RNA, Small Interfering
Figure
Reference
-
1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114:97–109.2. Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012; 107:359–364.3. Tate MC, Aghi MK. Biology of angiogenesis and invasion in glioma. Neurotherapeutics. 2009; 6:447–457.4. Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014; 1840:1063–1071.5. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010; 39:925–938.6. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013; 14:699–712.7. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009; 10:155–159.8. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013; 20:300–307.9. Zhang XQ, Leung GK. Long non-coding RNAs in glioma: functional roles and clinical perspectives. Neurochem Int. 2014; 77:78–85.10. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009; 106:11667–11672.11. Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl). 2013; 91:791–801.12. Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015; 290:3925–3935.13. Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014; 35:2731–2739.14. Liu WT, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL, Huang JF. LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol. 2014; 20:6314–6321.15. Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015; 36:2403–2407.16. Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015; 1852:166–174.17. Jiang Y, Li Y, Fang S, Jiang B, Qin C, Xie P, Zhou G, Li G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol Lett. 2014; 7:2135–2141.18. Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res. 2015; 13:98–106.19. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041.20. Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189.21. Xu Z, Zeng X, Tian D, Xu H, Cai Q, Wang J, Chen Q. MicroRNA-383 inhibits anchorage-independent growth and induces cell cycle arrest of glioma cells by targeting CCND1. Biochem Biophys Res Commun. 2014; 453:833–838.22. Annibali D, Whitfield JR, Favuzzi E, Jauset T, Serrano E, Cuartas I, Redondo-Campos S, Folch G, Gonzàlez-Juncà A, Sodir NM, et al. Myc inhibition is effective against glioma and reveals a role for Myc in proficient mitosis. Nat Commun. 2014; 5:4632.23. Qin X, Yao J, Geng P, Fu X, Xue J, Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int J Clin Exp Pathol. 2014; 7:3065–3072.24. Zhen L, Yun-Hui L, Hong-Yu D, Jun M, Yi-Long Y. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. Forthcoming. 2015.25. Zhang XQ, Kiang KM, Wang YC, Pu JK, Ho A, Cheng SY, Lee D, Zhang PD, Chen JJ, Lui WM, et al. IDH1 mutation-associated long non-coding RNA expression profile changes in glioma. J Neurooncol. 2015; 125:253–263.26. Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J, Li Z, Liu XB, Li ZQ, Wang ZH, et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015; 6:21934–21949.27. Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014; 68:557–564.28. Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015; 34:7.29. Lu H, He Y, Lin L, Qi Z, Ma L, Li L, Su Y. Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+ cervical cancer via sponging miR-145. Tumour Biol. Forthcoming. 2015.30. Guo F, Yu F, Wang J, Li Y, Li Y, Li Z, Zhou Q. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep. 2015; 3:309–312.31. Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015; 8:5395–5402.32. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012; 8:2289–2294.33. Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015; 36:3355–3359.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Long Noncoding RNA MALAT1 Regulates Hepatocellular Carcinoma Growth Under Hypoxia via Sponging MicroRNA-200a
- Roles of Long Non-Coding RNAs on Tumorigenesis and Glioma Development
- A Novel Type of Non-coding RNA, nc886, Implicated in Tumor Sensing and Suppression
- Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings
- Long Noncoding RNA PVT1 Promotes Stemness and Temozolomide Resistance through miR-365/ELF4/SOX2 Axis in Glioma