Yonsei Med J.
2019 Aug;60(8):727-734. 10.3349/ymj.2019.60.8.727.
Long Noncoding RNA MALAT1 Regulates Hepatocellular Carcinoma Growth Under Hypoxia via Sponging MicroRNA-200a
- Affiliations
-
- 1Infection Department, The First Hospital of Lanzhou University, Lanzhou, China.
- 2Department of Ultrasound, The First Hospital of Lanzhou University, Lanzhou, China. ldyyzhao@163.com
- KMID: 2452953
- DOI: http://doi.org/10.3349/ymj.2019.60.8.727
Abstract
- PURPOSE
Hepatocellular carcinoma (HCC) is a common cancer worldwide. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a long noncoding RNA (lncRNA), has been reported to be aberrantly expressed in hypoxic cancer cells. MALAT1 plays a significant role in many malignancies, including HCC. The aim of this study was to explore the role of MALAT1 in hypoxic HCC cells and its underlying regulatory mechanism.
MATERIALS AND METHODS
Quantitative reverse transcription PCR (qRT-PCR) assay was performed to detect the mRNA levels of MALAT1 and microRNA-200a (miR-200a) in HCC cells. Cell invasion and migration ability were evaluated by Transwell assay. Starbase v2.0 and luciferase reporter assay were employed to identify the association between MALAT1 and miR-200a. Cell proliferation and apoptosis were measured by MTT assay and flow cytometry, respectively.
RESULTS
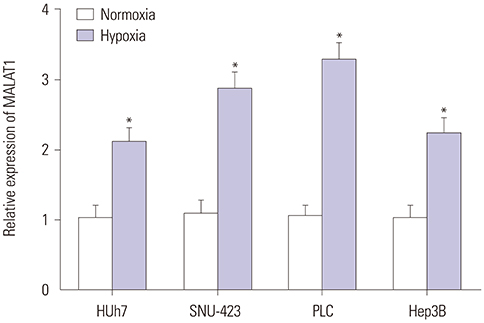
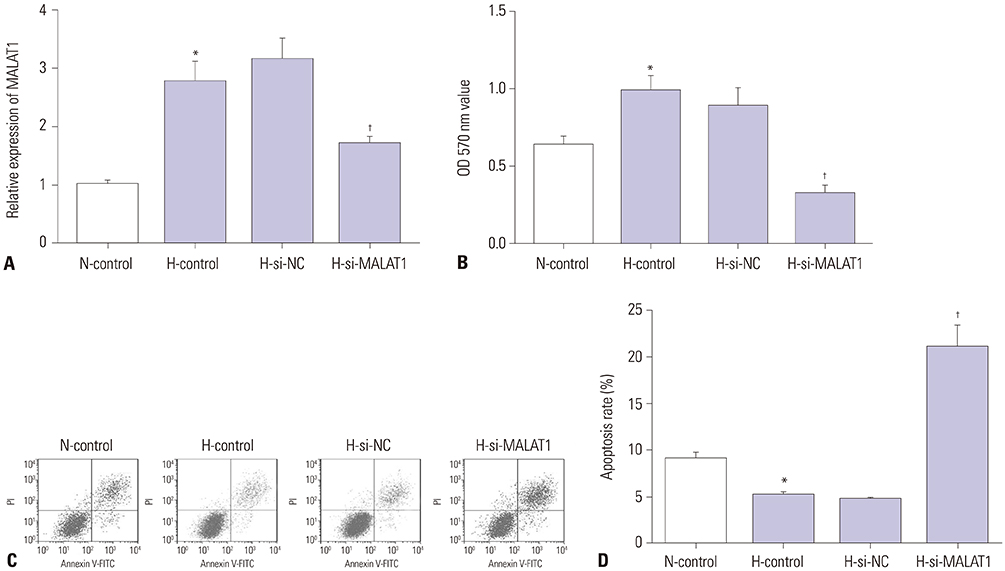
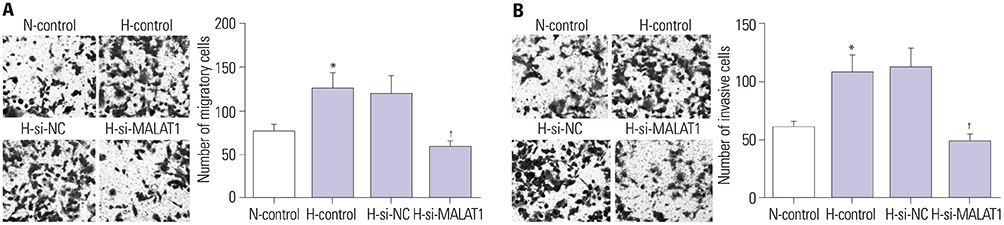
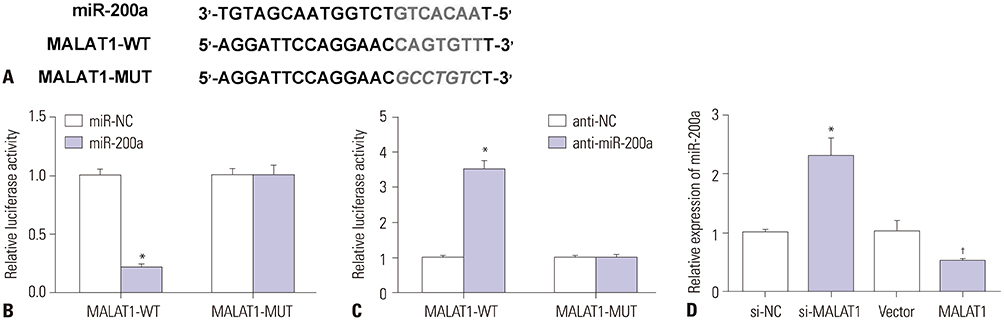
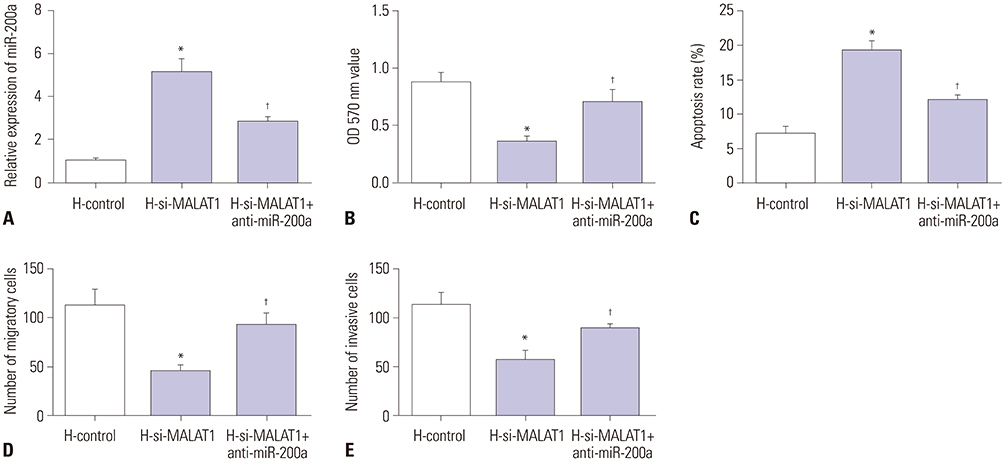
MALAT1 levels were significantly upregulated in HCC cells under hypoxia. Hypoxia promoted proliferation, migration, and invasion, and blocked apoptosis in Hep3B cells, which were weakened by knockdown of MALAT1. Starbase v2.0 showed that MALAT1 and miR-200a have a complementarity region, and luciferase reporter assay verified that MALAT1 interacted with miR-200a in Hep3B cells. Moreover, MALAT1 negatively regulated the expression of miR-200a. miR-200a levels were dramatically downregulated in HCC cells under hypoxia. Upregulation of miR-200a inhibited proliferation, migration, and invasion, and induced apoptosis in Hep3B cells under hypoxia. Interestingly, downregulation of miR-200a partially reversed the tumor-suppressive effect of knockdown of MALAT1 on Hep3B cells in hypoxic condition.
CONCLUSION
LncRNA MALAT1 was involved in proliferation, migration, invasion, and apoptosis by interacting with miR-200a in hypoxic Hep3B cells, revealing a new mechanism of MALAT1 involved in hypoxic HCC progression.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Clinical significance of exosomal noncoding RNAs in hepatocellular carcinoma: a narrative review
Jae Sung Yoo, Min Kyu Kang
J Yeungnam Med Sci. 2024;42:4. doi: 10.12701/jyms.2023.01186.
Reference
-
1. Ferenci P, Fried M, Labrecque D, Bruix J, Sherman M, Omata M, et al. Hepatocellular carcinoma (HCC): a global perspective. J Clin Gastroenterol. 2010; 44:239–245.2. Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010; 15:Suppl 4. 5–13.
Article3. Stuart KE, Anand AJ, Jenkins RL. Hepatocellular carcinoma in the United States. Prognostic features, treatment outcome, and survival. Cancer. 1996; 77:2217–2222.
Article4. Höckel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001; 93:266–276.
Article5. Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KS, et al. Hypoxia: importance in tumor biology, noninvasive measurement by imaging, and value of its measurement in the management of cancer therapy. Int J Radiat Biol. 2006; 82:699–757.
Article6. Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010; 1799:597–615.
Article7. Liu X, Wang Y, Sun L, Min J, Liu J, Chen D, et al. Long noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018; 109:988–1000.
Article8. Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018; 37:5811–5828.
Article9. Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open. 2015; 5:e008653.
Article10. Tian W, Du Y, Ma Y, Gu L, Zhou J, Deng D. MALAT1-miR663a negative feedback loop in colon cancer cell functions through direct miRNA-lncRNA binding. Cell Death Dis. 2018; 9:857.
Article11. Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015; 290:3925–3935.
Article12. Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015; 1852:166–174.
Article13. Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017; 39:1010428317718135.
Article14. Mehta A, Herrera H, Block T. Glycosylation and liver cancer. Adv Cancer Res. 2015; 126:257–279.
Article15. Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016; 2:16018.
Article16. Shi B, Zhang X, Chao L, Zheng Y, Tan Y, Wang L, et al. Comprehensive analysis of key genes, microRNAs and long non-coding RNAs in hepatocellular carcinoma. FEBS Open Bio. 2018; 8:1424–1436.
Article17. Wu Y, Huang C, Meng X, Li J. Long noncoding RNA MALAT1: insights into its biogenesis and implications in human disease. Curr Pharm Des. 2015; 21:5017–5028.
Article18. Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS, et al. Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother. 2018; 102:653–669.
Article19. Zhao M, Wang S, Li Q, Ji Q, Guo P, Liu X. MALAT1: a long non-coding RNA highly associated with human cancers. Oncol Lett. 2018; 16:19–26.20. Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017; 77:1155–1167.
Article21. Chen L, Yao H, Wang K, Liu X. Long non-coding RNA MALAT1 regulates ZEB1 expression by sponging miR-143-3p and promotes hepatocellular carcinoma progression. J Cell Biochem. 2017; 118:4836–4843.
Article22. Guil S, Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci. 2015; 40:248–256.
Article23. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016; 96:1297–1325.
Article24. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005; 122:6–7.
Article25. Chu R, Mo G, Duan Z, Huang M, Chang J, Li X, et al. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun Signal. 2014; 12:45.
Article26. Song Y, Wang F, Huang Q, Cao Y, Zhao Y, Yang C. MicroRNAs contribute to hepatocellular carcinoma. Mini Rev Med Chem. 2015; 15:459–466.
Article27. Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018; 18:116.
Article28. Tak H, Kang H, Ji E, Hong Y, Kim W, Lee EK. Potential use of TIA-1, MFF, microRNA-200a-3p, and microRNA-27 as a novel marker for hepatocellular carcinoma. Biochem Biophys Res Commun. 2018; 497:1117–1122.
Article29. Chen SY, Ma DN, Chen QD, Zhang JJ, Tian YR, Wang ZC, et al. MicroRNA-200a inhibits cell growth and metastasis by targeting Foxa2 in hepatocellular carcinoma. J Cancer. 2017; 8:617–625.
Article30. Yang X, Wang J, Qu S, Zhang H, Ruan B, Gao Y, et al. MicroRNA-200a suppresses metastatic potential of side population cells in human hepatocellular carcinoma by decreasing ZEB2. Oncotarget. 2015; 6:7918–7929.
Article31. Li Y, Zhu X, Liu X, Du A, Yu B. MiR-200a mediates protection of thymosin beta 4 in cardiac microvascular endothelial cells as a novel mechanism under hypoxia-reoxygenation injury. . J Cell Biochem. 2019; 07. 02. DOI: 10.1002/jcb.29237. [Epub].32. Sun X, Zuo H, Liu C, Yang Y. Overexpression of miR-200a protects cardiomyocytes against hypoxia-induced apoptosis by modulating the kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 signaling axis. Int J Mol Med. 2016; 38:1303–1311.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Silencing of Long Non-Coding RNA MALAT1 Promotes Apoptosis of Glioma Cells
- XIST Induced by JPX Suppresses Hepatocellular Carcinoma by Sponging miR-155-5p
- LncRNA MALAT1/MiR-145 Adjusts IL-1β-Induced Chondrocytes Viability and Cartilage Matrix Degradation by Regulating ADAMTS5 in Human Osteoarthritis
- Clinical significance of exosomal noncoding RNAs in hepatocellular carcinoma: a narrative review
- Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings