Cancer Res Treat.
2007 Sep;39(3):116-124.
Functional Link between DNA Damage Responses and Transcriptional Regulation by ATM in Response to a Histone Deacetylase Inhibitor TSA
- Affiliations
-
- 1Department of Biological Sciences, College of Natural Sciences and Department of Molecular Science and Technology, Ajou University, Suwon, Korea. jsjlee@ajou.ac.kr
Abstract
-
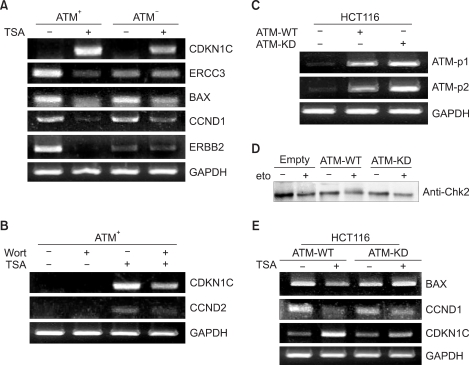
PURPOSE: Mutations in the ATM (ataxia-telangiectasia mutated) gene, which encodes a 370 kd protein with a kinase catalytic domain, predisposes people to cancers, and these mutations are also linked to ataxia-telangiectasia (A-T). The histone acetylaion/deacetylation- dependent chromatin remodeling can activate the ATM kinase-mediated DNA damage signal pathway (in an accompanying work, Lee, 2007). This has led us to study whether this modification can impinge on the ATM-mediated DNA damage response via transcriptional modulation in order to understand the function of ATM in the regulation of gene transcription.
MATERIALS AND METHODS
To identify the genes whose expression is regulated by ATM in response to histone deaceylase (HDAC) inhibition, we performed an analysis of oligonucleotide microarrays with using the appropriate cell lines, isogenic A-T (ATM square) and control (ATM+) cells, following treatment with a HDAC inhibitor TSA.
RESULTS
Treatment with TSA reprograms the differential gene expression profile in response to HDAC inhibition in ATM square cells and ATM+ cells. We analyzed the genes that are regulated by TSA in the ATM-dependent manner, and we classified these genes into different functional categories, including those involved in cell cycle/DNA replication, DNA repair, apoptosis, growth/differentiation, cell- cell adhesion, signal transduction, metabolism and transcription.
CONCLUSION
We found that while some genes are regulated by TSA without regard to ATM, the patterns of gene regulation are differentially regulated in an ATM-dependent manner. Taken together, these finding indicate that ATM can regulate the transcription of genes that play critical roles in the molecular response to DNA damage, and this response is modulated through an altered HDAC inhibition-mediated gene expression.
Keyword
MeSH Terms
-
Apoptosis
Ataxia Telangiectasia
Catalytic Domain
Cell Adhesion
Cell Line
Chromatin Assembly and Disassembly
DNA Damage*
DNA Replication
DNA*
Gene Expression
Histone Deacetylase Inhibitors*
Histone Deacetylases*
Histones*
Metabolism
Oligonucleotide Array Sequence Analysis
Phosphotransferases
Signal Transduction
Transcriptome
DNA
Histone Deacetylase Inhibitors
Histone Deacetylases
Histones
Phosphotransferases
Figure
Reference
-
1. Kastan MB, Lim DS. The many substrates and functions of ATM. Nat Rev Mol Cell Biol. 2000; 1:179–186. PMID: 11252893.
Article2. Jang ER, Lee JH, Lim DS, Lee JS. Analysis of ataxia-telangiectasia mutated (ATM)- and Nijmegen breakage syndrome (NBS)-regulated gene expression patterns. J Cancer Res Clin Oncol. 2004; 130:225–234. PMID: 14745549.
Article3. Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, et al. ATM-depedent phosphoryaltion of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001; 15:1067–1077. PMID: 11331603.4. Welcsh PL, Lee MK, Gonzalez-Hernandez RM, Black DJ, Mahadevappa M, Swisher EM, et al. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci U S A. 2002; 99:7560–7565. PMID: 12032322.
Article5. Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol. 2004; 24:976–984. PMID: 14729946.
Article6. Foray N, Marot D, Gabriel A, Randrianarison V, Carr AM, Perricaudet M, et al. A subset of ATM- and ATR-dependent phsophorylation events requires the BRCA1 protein. EMBO J. 2003; 22:2860–2871. PMID: 12773400.7. Wu-Baer F, Baer R. Effect of DNA damage on a BRCA1 complex. Nature. 2001; 414:36. PMID: 11689934.
Article8. Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRb at ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 2007; 26:2083–2093. PMID: 17380128.
Article9. Bruno T, De Nicola F, Iezzi S, Lecis D, D'Angelo C, Di Padova M, et al. Che-1 phosphorylation by ATM/ATR and Chk2-kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell. 2006; 10:473–486. PMID: 17157788.10. Glozak MA, Seto E. Histone deacetylase and cancer. Oncogene. 2007; 26:5420–5432. PMID: 17694083.11. Chen CS, Wang YC, Yang HC, Huang PH, Kulp SK, Yang CC, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agent that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007; 67:5318–5327. PMID: 17545612.12. Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998; 18:7185–7191. PMID: 9819405.
Article13. Noh EJ, Lee JS. Functional interplay between modulation of histone deacetylase activity and its regulatory role in G2-M transition. Biochem Biophys Res Commun. 2003; 310:267–273. PMID: 14521905.
Article14. Jang ER, JS Lee. Activation of ATM-dependent DNA damage signal pathway by a histone deacetylase inhibitor, Trichostatin A. Cancer Res Treat. 2007; Submitted.
Article15. Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003; 421:499–506. PMID: 12556884.
Article16. Vergani L, Fugazza G, Chessa L, Nicolini C. Changes of chromatin condensation in one patient with ataxia telangiectasia disorder: a structural study. J Cell Biochem. 1999; 75:578–586. PMID: 10572241.
Article17. Pedeux R, Lefort K, Cuenin C, Cortes U, Kellner K, Dore JF, et al. Specific induction of gadd45 in human melanocytes and melanoma cells after UVB irradiation. Int J Cancer. 2002; 98:811–816. PMID: 11948456.18. Cheng L, Guan Y, Li L, Legerski RJ, Einspahr J, Bangert J, et al. Expression in normal human tissues of five nucleotide excision repair genes measured simultaneously by multiplex reverse transcriptase-polymerase chain reaction. Cancer Epidemiol Biomarkers Prev. 1999; 8:801–807. PMID: 10498399.19. Ayesh S, Matouk I, Schneider T, Ohana P, Laster M, Al-Sharef W, et al. Possible physiological role of H19 RNA. Mol Carcinog. 2002; 35:63–74. PMID: 12325036.
Article20. Yu J, Miehlke S, Ebert MP, Szokodi D, Wehvnignh B, Malfertheiner P, et al. Expression of cyclin genes in human gastric cancer and in first degree relatives. Chin Med J (Engl). 2002; 115:710–715. PMID: 12133540.21. Ejima Y, Yang L, Sasaki MS. Aberrant splicing of the ATM gene associated with shortening of the intronic mononucleotide tract in human colon tumor cell lines: a novel mutation target of microsatellite instability. Int J Cancer. 2000; 86:262–268. PMID: 10738255.22. Harter L, Mica L, Stocker R, Trentz O, Keel M. Mcl-1 correlates with reduced apoptosis in neutrophils from patients with sepsis. J Am Coll Surg. 2003; 197:964–973. PMID: 14644285.23. Thallinger C, Wolschek MF, Wacheck V, Maierhofer H, Gunsberg P, Polterauer P, et al. Mcl-1 antisense therapy chemosensitizes human melanoma in a SCID mouse xenotransplantation model. J Invest Dermatol. 2003; 120:1081–1086. PMID: 12787138.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Activation of ATM-dependent DNA Damage Signal Pathway by a Histone Deacetylase Inhibitor, Trichostatin A
- ATM modulates transcription in response to histone deacetylase inhibition as part of its DNA damage response
- Coordinated transcriptional regulation of calmegin, a testis-specific molecular chaperon, by histone deacetylase and CpG methyltransferase
- Effects of Trichostatin A on the Chondrogenesis from Human Mesenchymal Stem Cells
- Trichostatin A Protects Liver against Septic Injury through Inhibiting Toll-Like Receptor Signaling