J Bacteriol Virol.
2012 Sep;42(3):211-223. 10.4167/jbv.2012.42.3.211.
Proteomic Analysis of Thiol-active Proteins of Helicobacter pylori 26695
- Affiliations
-
- 1Department of Microbiology, Gyeongsang National University College of Medicine, Jinju, Gyeong-Nam, Korea. mjecho@gnu.kr
- 2Department of Pediatrics, Gyeongsang National University College of Medicine, Jinju, Gyeong-Nam, Korea.
- 3Research Institute of Life Science, Gyeongsang National University, Jinju, Gyeong-Nam, Korea.
- KMID: 1434750
- DOI: http://doi.org/10.4167/jbv.2012.42.3.211
Abstract
- Helicobacter pylori are a capnophilic bacterium, which colonize gastric mucosa and are resistant to acidic and oxidative damage. Thiol-active proteins subserve redox functions in tolerating oxidative stress and environmental toxicants, such as hydrogen peroxide and hypochlorous acid. We analyzed disulfide-containing proteins of H. pylori strain 26695. Active disulfide-containing proteins were separated by thiol-affinity chromatography, displayed with two-dimensional electrophoresis (2-DE), and identified by MALDI-TOF-MS. Thirty-five putative disulfide proteins, including AhpC (HP1563), GroEL (HP0011), and FrdB (HP0191), were identified in this study. In addition, 4 disulfide proteins of HypB, FusA, TufB, and AhpC showed enhanced intensities in the periplasmic space when compared with the pellet, suggesting that these proteins might play roles in the first redox system against environmental oxidative stresses. Disulfide-containing proteins identified in this study will provide the standard landscape for constructing the proteome components responsible for redox regulation of H. pylori.
Keyword
MeSH Terms
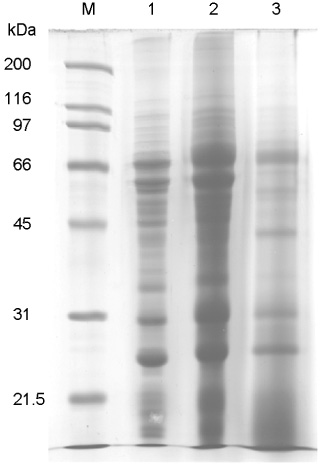
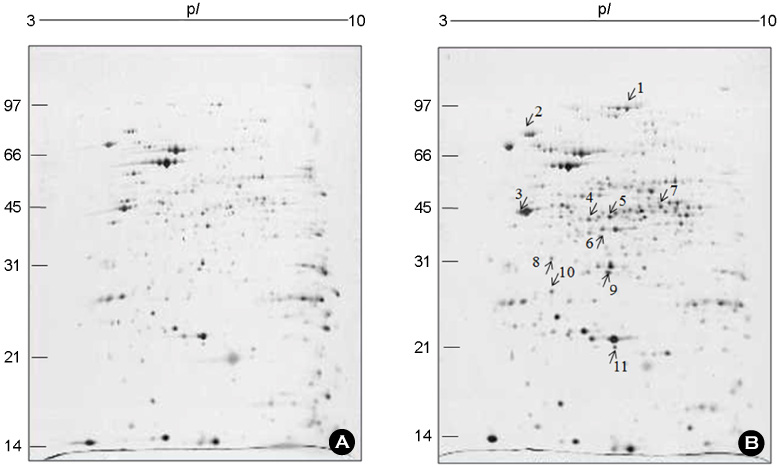
Figure
Cited by 1 articles
-
Proteome Analysis of Alkylhydroxide Peroxidase-Deficient Isogenic Mutant of Helicobacter pylori 26695
Woo-Kon Lee, Seung-Chul Baik, Min-Kyung Shin, Myunghwan Jung, Jin-Sik Park, Jong-Hoon Ha, Dong-Hae Lee, Min-Jeong Kim, Jeong-ih Shin, Hyung-Lyun Kang
J Bacteriol Virol. 2019;49(4):191-202. doi: 10.4167/jbv.2019.49.4.191.
Reference
-
1. Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984. 1:1311–1315.
Article2. Graham DY, Malaty HM, Evans DG, Evans DJ Jr, Klein PD, Adam E. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Effect of age, race, and socioeconomic status. Gastroenterology. 1991. 100:1495–1501.
Article3. Wolle K, Leodolter A, Malfertheiner P, König W. Antibiotic susceptibility of Helicobacter pylori in Germany: stable primary resistance from 1995 to 2000. J Med Microbiol. 2002. 51:705–709.
Article4. Aktay AN, Werlin SL, Hellman RS. The impact of solid-phase gastric emptying studies in the management of children with dyspepsia. Clin Pediatr (Phila). 2003. 42:621–625.
Article5. Dooley CP, Cohen H, Fitzgibbons PL, Bauer M, Appleman MD, Perez-Perez GI, et al. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med. 1989. 321:1562–1566.
Article6. Blaser MJ. Gastric Campylobacter-like organisms, gastritis, and peptic ulcer disease. Gastroenterology. 1987. 93:371–383.
Article7. Sidebotham RL, Baron JH. Hypothesis: Helicobacter pylori, urease, mucus, and gastric ulcer. Lancet. 1990. 335:193–195.8. Mégraud F, Lamouliatte H. Helicobacter pylori and duodenal ulcer. Evidence suggesting causation. Dig Dis Sci. 1992. 37:769–772.9. Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, Ko GH, et al. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996. 56:1279–1282.10. Forman D, Newell DG, Fullerton F, Yarnell JW, Stacey AR, Wald N, et al. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. BMJ. 1991. 302:1302–1305.
Article11. Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991. 325:1132–1136.
Article12. Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991. 325:1127–1131.
Article13. Mendz GL, Jimenez BM, Hazell SL, Gero AM, O'Sullivan WJ. De novo synthesis of pyrimidine nucleotides by Helicobacter pylori. J Appl Bacteriol. 1994. 77:1–8.14. Burns BP, Hazell SL, Mendz GL. Acetyl-CoA carboxylase activity in Helicobacter pylori and the requirement of increased CO2 for growth. Microbiology. 1995. 141:3113–3118.
Article15. Joo JS, Park KC, Song JY, Kim DH, Lee KJ, Kwon YC, et al. A thin-layer liquid culture technique for the growth of Helicobacter pylori. Helicobacter. 2010. 15:295–302.
Article16. Kettenhofen NJ, Wood MJ. Formation, reactivity, and detection of protein sulfenic acids. Chem Res Toxicol. 2010. 23:1633–1646.
Article17. Leonard SE, Carroll KS. Chemical 'omics' approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011. 15:88–102.
Article18. Klomsiri C, Karplus PA, Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011. 14:1065–1077.
Article19. Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007. 7:381–391.
Article20. Sarajärvi T, Tuusa JT, Haapasalo A, Lackman JJ, Sormunen R, Helisalmi S, et al. Cysteine 27 variant of the delta-opioid receptor affects amyloid precursor protein processing through altered endocytic trafficking. Mol Cell Biol. 2011. 31:2326–2340.
Article21. Subramanian S, Sijwali PS, Rosenthal PJ. Falcipain cysteine proteases require bipartite motifs for trafficking to the Plasmodium falciparum food vacuole. J Biol Chem. 2007. 282:24961–24969.
Article22. Guttmann RP. Redox regulation of cysteine-dependent enzymes. J Anim Sci. 2010. 88:1297–1306.23. Spadaro D, Yun BW, Spoel SH, Chu C, Wang YQ, Loake GJ. The redox switch : dynamic regulation of protein function by cysteine modifications. Physiol Plant. 2010. 138:360–371.
Article24. Cross JV, Templeton DJ. Regulation of signal transduction through protein cysteine oxidation. Antioxid Redox Signal. 2006. 8:1819–1827.
Article25. Franklin CC, Backos DS, Mohar I, White CC, Forman HJ, Kavanagh TJ. Structure, function, and post-translational regulation of the catalytic and modifier subunits of glutamate cysteine ligase. Mol Aspects Med. 2009. 30:86–98.
Article26. Stipanuk MH, Ueki I, Dominy JE Jr, Simmons CR, Hirschberger LL. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids. 2009. 37:55–63.
Article27. Turell L, Botti H, Carballal S, Radi R, Alvarez B. Sulfenic acid--a key intermediate in albumin thiol oxidation. J Chromatogr B Analyt Technol Biomed Life Sci. 2009. 877:3384–3392.
Article28. Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991. 67:581–589.
Article29. Gleiter S, Bardwell JC. Disulfide bond isomerization in prokaryotes. Biochim Biophys Acta. 2008. 1783:530–534.
Article30. Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008. 1783:549–556.
Article31. Inaba K. Structural basis of protein disulfide bond generation in the cell. Genes Cells. 2010. 15:935–943.32. Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006. 31:455–464.
Article33. Okazaki S, Tachibana T, Naganuma A, Mano N, Kuge S. Multistep disulfide bond formation in Yap1 is required for sensing and transduction of H2O2 stress signal. Mol Cell. 2007. 27:675–688.
Article34. O'Brian CA, Chu F. Post-translational disulfide modifications in cell signaling--role of inter-protein, intra-protein, S-glutathionyl, and S-cysteaminyl disulfide modifications in signal transmission. Free Radic Res. 2005. 39:471–480.35. Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009. 394:147–158.
Article36. Lindahl M, Mata-Cabana A, Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal. 2011. 14:2581–2642.
Article37. Kumar JK, Tabor S, Richardson CC. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2004. 101:3759–3764.
Article38. Pérez-Pérez ME, Florencio FJ, Lindahl M. Selecting thioredoxins for disulphide proteomics: target proteomes of three thioredoxins from the cyanobacterium Synechocystis sp. PCC 6803. Proteomics. 2006. 6:S186–S195.39. Hochgräfe F, Mostertz J, Albrecht D, Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol Microbiol. 2005. 58:409–425.
Article40. Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004. 2:e333.41. Hu W, Tedesco S, McDonagh B, Bárcena JA, Keane C, Sheehan D. Selection of thiol- and disulfide-containing proteins of Escherichia coli on activated thiol-sepharose. Anal Biochem. 2010. 398:245–253.
Article42. Brandes N, Rinck A, Leichert LI, Jakob U. Nitrosative stress treatment of E. coli targets distinct set of thiol-containing proteins. Mol Microbiol. 2007. 66:901–914.
Article43. Charles RL, Schröder E, May G, Free P, Gaffney PR, Wait R, et al. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007. 6:1473–1484.44. Yano H, Kuroda S. Introduction of the disulfide proteome: application of a technique for the analysis of plant storage proteins as well as allergens. J Proteome Res. 2008. 7:3071–3079.
Article45. Yang Y, Song Y, Loscalzo J. Regulation of the protein disulfide proteome by mitochondria in mammalian cells. Proc Natl Acad Sci U S A. 2007. 104:10813–10817.
Article46. García-Santamarina S, Boronat S, Espadas G, Ayté J, Molina H, Hidalgo E. The oxidized thiol proteome in fission yeast--optimization of an ICAT-based method to identify H2O2-oxidized proteins. J Proteomics. 2011. 74:2476–2486.
Article47. McDonagh B, Padilla CA, Pedrajas JR, Bárcena JA. Biosynthetic and iron metabolism is regulated by thiol proteome changes dependent on glutaredoxin-2 and mitochondrial peroxiredoxin-1 in Saccharomyces cerevisiae. J Biol Chem. 2011. 286:15565–15576.
Article48. Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010. 468:790–795.
Article49. McDonagh B, Sheehan D. Effect of oxidative stress on protein thiols in the blue mussel Mytilus edulis: proteomic identification of target proteins. Proteomics. 2007. 7:3395–3403.
Article50. Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, et al. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006. 5:361–369.
Article51. Liu T, Qian WJ, Chen WN, Jacobs JM, Moore RJ, Anderson DJ, et al. Improved proteome coverage by using high efficiency cysteinyl peptide enrichment: the human mammary epithelial cell proteome. Proteomics. 2005. 5:1263–1273.
Article52. Song JY, Choi YJ, Kim JM, Kim YR, Jo JS, Park JS, et al. Purification and characterization of Helicobacter pylori γ-glutamyltranspeptidase. J Bacteriol Virol. 2011. 41:255–265.
Article53. Lee K, Lee J, Kim Y, Bae D, Kang KY, Yoon SC, et al. Defining the plant disulfide proteome. Electrophoresis. 2004. 25:532–541.
Article54. Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003. 49:219–234.
Article55. Park JW, Lee SG, Song JY, Jun JS, Joo JS, Youn HS, et al. Proteomic analysis of Helicoacter pylori whole cell proteins using the narrow range IPG strips. J Bacteriol Virol. 2007. 37:203–212.
Article56. Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999. 20:601–605.
Article57. O'Connell KL, Stults JT. Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis. 1997. 18:349–359.58. Harder A, Wildgruber R, Nawrocki A, Fey SJ, Larsen PM, Görg A. Comparison of yeast cell protein solubilization procedures for two-dimensional electrophoresis. Electrophoresis. 1999. 20:826–829.
Article59. Nandakumar MP, Marten MR. Comparison of lysis methods and preparation protocols for one- and two-dimensional electrophoresis of Aspergillus oryzae intracellular proteins. Electrophoresis. 2002. 23:2216–2222.
Article60. Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006. 61:847–860.
Article61. Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci U S A. 2006. 103:2552–2557.
Article62. Comtois SL, Gidley MD, Kelly DJ. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology. 2003. 149:121–129.
Article63. Windle HJ, Fox A, Ní Eidhin D, Kelleher D. The thioredoxin system of Helicobacter pylori. J Biol Chem. 2000. 275:5081–5089.64. Schauer K, Muller C, Carrière M, Labigne A, Cavazza C, De Reuse H. The Helicobacter pylori GroES cochaperonin HspA functions as a specialized nickel chaperone and sequestration protein through its unique C-terminal extension. J Bacteriol. 2010. 192:1231–1237.
Article65. Radford SE. GroEL: More than Just a folding cage. Cell. 2006. 125:831–833.
Article66. Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat Struct Biol. 2001. 8:505–509.67. Pinkse MW, Maier CS, Kim JI, Oh BH, Heck AJ. Macromolecular assembly of Helicobacter pylori urease investigated by mass spectrometry. J Mass Spectrom. 2003. 38:315–320.
Article68. Mittl PR, Lüthy L, Hunziker P, Grütter MG. The cysteine-rich protein A from Helicobacter pylori is a beta-lactamase. J Biol Chem. 2000. 275:17693–17699.69. Ogura M, Perez JC, Mittl PR, Lee HK, Dailide G, Tan S, et al. Helicobacter pylori evolution: lineage-specific adaptations in homologs of eukaryotic Sel1-like genes. PLoS Comput Biol. 2007. 3:e151.70. Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004. 53:1397–1406.
Article71. Sardesai AA, Genevaux P, Schwager F, Ang D, Georgopoulos C. The OmpL porin does not modulate redox potential in the periplasmic space of Escherichia coli. EMBO J. 2003. 22:1461–1466.
Article72. Genevaux P, Bauda P, DuBow MS, Oudega B. Identification of Tn10 insertions in the dsbA gene affecting Escherichia coli biofilm formation. FEMS Microbiol Lett. 1999. 173:403–409.
Article73. Cho MJ, Jeon BS, Park JW, Jung TS, Song JY, Lee WK, et al. Identifying the major proteome components of Helicobacter pylori strain 26695. Electrophoresis. 2002. 23:1161–1173.
Article74. Lee HW, Choe YH, Kim DK, Jung SY, Lee NG. Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori: regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron. Proteomics. 2004. 4:2014–2027.
Article75. Backert S, Kwok T, Schmid M, Selbach M, Moese S, Peek RM Jr, et al. Subproteomes of soluble and structure-bound Helicobacter pylori proteins analyzed by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2005. 5:1331–1345.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Proteome Analysis of Alkylhydroxide Peroxidase-Deficient Isogenic Mutant of Helicobacter pylori 26695
- Sequence Analysis of Hypothetical Proteins from Helicobacter pylori 26695 to Identify Potential Virulence Factors
- Proteomic Analysis of Helicobacter pylori Whole Cell Proteins using the Narrow Range IPG Strips
- Treatment of Helicobacter pylori infection in functional dyspepsia
- Application of Hemin-Agarose Affinity Chromatography to Enrich Proteome Components of Helicobacter pylori Strain 26695