J Gastric Cancer.
2019 Dec;19(4):460-472. 10.5230/jgc.2019.19.e43.
LINC00703 Acts as a Tumor Suppressor via Regulating miR-181a/KLF6 Axis in Gastric Cancer
- Affiliations
-
- 1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China. zj.qian@siat.ac.cn
- 2University of Chinese Academy of Sciences, Beijing, China.
- 3Department of Pathophysiology, Shenzhen University, Shenzhen, China.
- 4East Hospital Affiliated to Tongji University, Shanghai, China.
- KMID: 2466034
- DOI: http://doi.org/10.5230/jgc.2019.19.e43
Abstract
- PURPOSE
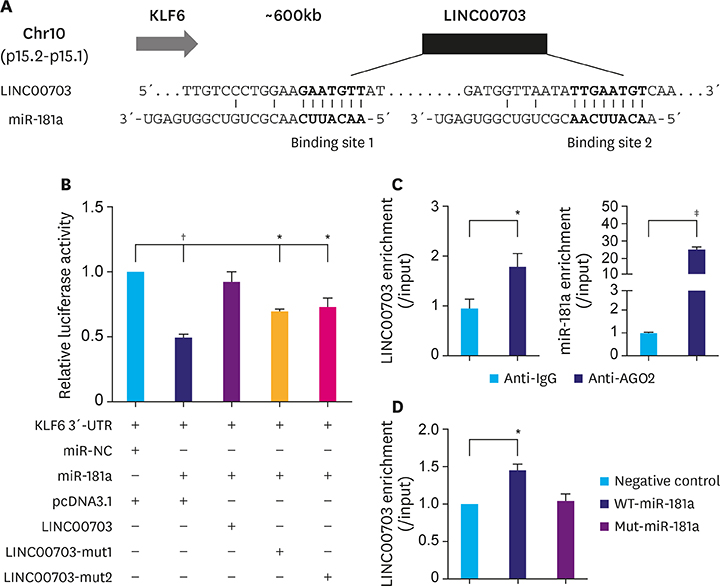
Long noncoding RNA 00703 (LINC00703) was found originating from a region downstream of Kruppel-like factor 6 (KLF6) gene, having 2 binding sites for miR-181a. Since KLF6 has been reported as a target of miR-181a in gastric cancer (GC), this study aims to investigate whether LINC00703 regulates the miR-181a/KLF6 axis and plays a functional role in GC pathogenesis.
MATERIALS AND METHODS
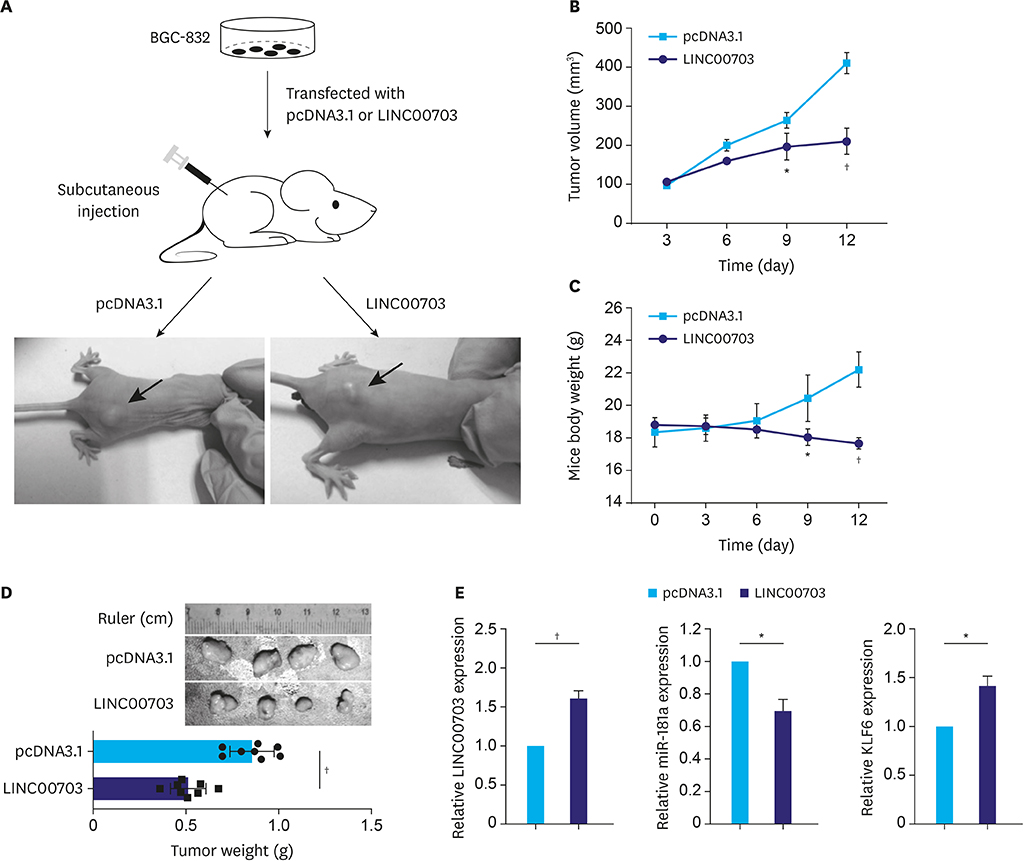
GC tissues, cell lines, and nude mice were included in this study. RNA binding protein immunoprecipitation (RIP) and pull-down assays were used to evaluate interaction between LINC00703 and miR-181a. Quantitative real-time polymerase chain reaction and western blot were applied for analysis of gene expression at the transcriptional and protein levels. A nude xenograft mouse model was used to determine LINC00703 function in vivo.
RESULTS
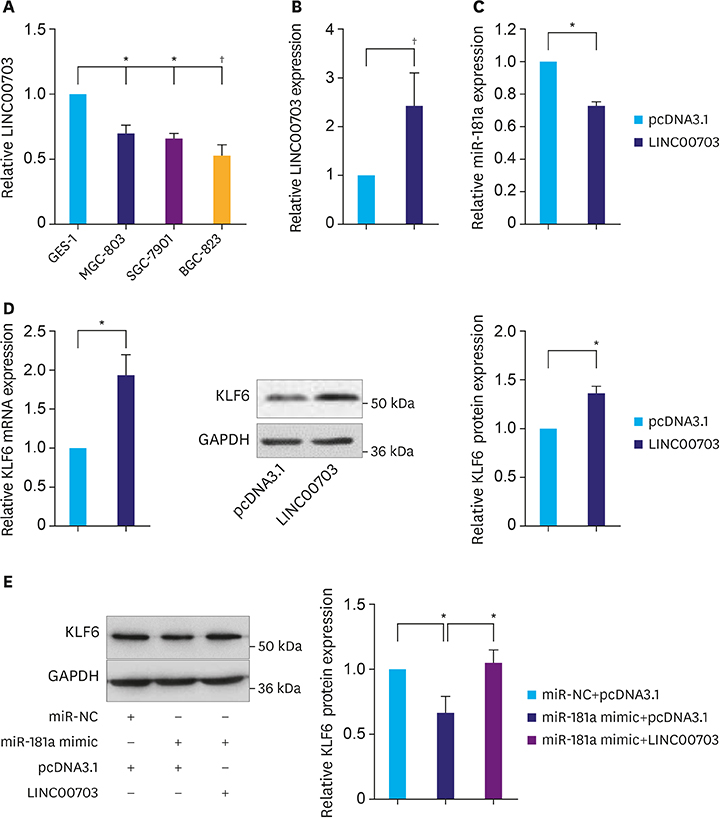
We revealed that LINC00703 competitively interacts with miR-181a to regulate KLF6. Overexpression of LINC00703 inhibited cell proliferation, migration/invasion, but promoted apoptosis in vitro, and arrested tumor growth in vivo. LINC00703 expression was found to be decreased in GC tissues, which was positively correlated with KLF6, but negatively with the miR-181a levels.
CONCLUSIONS
LINC00703 may have an anti-cancer function via modulation of the miR-181a/KLF6 axis. This study also provides a new potential diagnostic marker and therapeutic target for GC treatment.
Keyword
MeSH Terms
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66:7–30.2. Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006; 12:354–362.3. Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009; 374:477–490.4. Matsunaga N, Wakasaki T, Yasumatsu R, Kotake Y. Long noncoding RNA, ANRIL, regulates the proliferation of head and neck squamous cell carcinoma. Anticancer Res. 2019; 39:4073–4077.5. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009; 136:629–641.6. Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013; 33:3185–3193.7. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009; 23:1494–1504.8. Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 2015; 36:3129–3136.9. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011; 147:358–369.10. Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014; 13:92.11. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumour Biol. 2016; 37:16345–16355.12. Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015; 464:807–813.13. Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015; 34:79.14. McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010; 90:1337–1381.15. Sangodkar J, Shi J, DiFeo A, Schwartz R, Bromberg R, Choudhri A, et al. Functional role of the KLF6 tumour suppressor gene in gastric cancer. Eur J Cancer. 2009; 45:666–676.16. Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005; 24:4588–4590.17. Bi J, Zeng X, Zhao L, Wei Q, Yu L, Wang X, et al. miR-181a induces macrophage polarized to M2 phenotype and promotes M2 macrophage-mediated tumor cell metastasis by targeting KLF6 and C/EBPα. Mol Ther Nucleic Acids. 2016; 5:e368.18. Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y, et al. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urol Oncol. 2018; 36:93.e23–93.e37.19. Zhang X, Nie Y, Du Y, Cao J, Shen B, Li Y. MicroRNA-181a promotes gastric cancer by negatively regulating tumor suppressor KLF6. Tumour Biol. 2012; 33:1589–1597.20. Liu H, Li J, Koirala P, Ding X, Chen B, Wang Y, et al. Long non-coding RNAs as prognostic markers in human breast cancer. Oncotarget. 2016; 7:20584–20596.21. Qian Z, Li Y, Yang H, Chen J, Li X, Gou D. PDGFBB promotes proliferation and migration via regulating miR-1181/STAT3 axis in human pulmonary arterial smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2018; 315:L965–L976.22. Li Y, Li L, Qian Z, Lin B, Chen J, Luo Y, et al. Phosphatidylinositol 3-kinase-DNA methyltransferase 1-miR-1281-histone deacetylase 4 regulatory axis mediates platelet-derived growth factor-induced proliferation and migration of pulmonary artery smooth muscle cells. J Am Heart Assoc. 2018; 7:e007572.23. Qian Z, Li Y, Chen J, Li X, Gou D. miR-4632 mediates PDGF-BB-induced proliferation and antiapoptosis of human pulmonary artery smooth muscle cells via targeting cJUN. Am J Physiol Cell Physiol. 2017; 313:C380–C391.24. Ma J, Yao Y, Wang P, Liu Y, Zhao L, Li Z, et al. MiR-181a regulates blood-tumor barrier permeability by targeting Krüppel-like factor 6. J Cereb Blood Flow Metab. 2014; 34:1826–1836.25. Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, et al. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017; 31:893–903.26. Zhao H, Peng R, Liu Q, Liu D, Du P, Yuan J, et al. The lncRNA H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016; 610:1–7.27. Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D, Quan ZW. Long noncoding RNA H19 contributes to gallbladder cancer cell proliferation by modulated miR-194-5p targeting AKT2. Tumour Biol. 2016; 37:9721–9730.28. Sun L, Sun P, Zhou QY, Gao X, Han Q. Long noncoding RNA MALAT1 promotes uveal melanoma cell growth and invasion by silencing of miR-140. Am J Transl Res. 2016; 8:3939–3946.29. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011; 472:120–124.30. Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012; 19:1068–1075.31. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011; 43:904–914.32. Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016; 33:88.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression Pattern of KLF6 in Korean Gastric Cancers
- MiR-181a Promotes Spermatogenesis by Targeting the S6K1 Pathway
- miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple Negative Breast Cancer
- MicroRNA-181a-5p Curbs Osteogenic Differentiation and Bone Formation Partially Through Impairing Runx1-Dependent Inhibition of AIF-1 Transcription
- The Role of Gastrokine 1 in Gastric Cancer