Ann Lab Med.
2020 Mar;40(2):155-163. 10.3343/alm.2020.40.2.155.
Upregulated Long Noncoding RNA LINC01234 Predicts Unfavorable Prognosis for Colorectal Cancer and Negatively Correlates With KLF6 Expression
- Affiliations
-
- 1Department of Ultrasound, Wuxi People's Hospital Affiliated to Nanjing Medical University, Wuxi, Jiangsu, China. zzqqyy88@163.com
- KMID: 2460913
- DOI: http://doi.org/10.3343/alm.2020.40.2.155
Abstract
- BACKGROUND
LINC01234, a long noncoding RNA (lncRNA), is overexpressed in several cancers, including colorectal cancer (CRC). We investigated the role of LINC01234 in CRC development and confirmed its correlation with Krüppel-like factor 6 (KLF6), a tumor suppressor gene that is dysregulated in CRC.
METHODS
We tested mRNA levels using quantitative reverse transcription PCR (qRT-PCR). Tissue samples from patients with CRC, inflammatory bowel disease (IBD), hyperplastic polyp, and adenoma were included. Correlations between clinicopathological parameters, overall survival (OS) rate, and LINC01234 were analyzed using Kruskal-Wallis H test. Additionally, cell proliferation, apoptosis, and tumor formation in nude mice were tested to investigate the mechanism of LINC01234. Western blotting was used to determine protein levels.
RESULTS
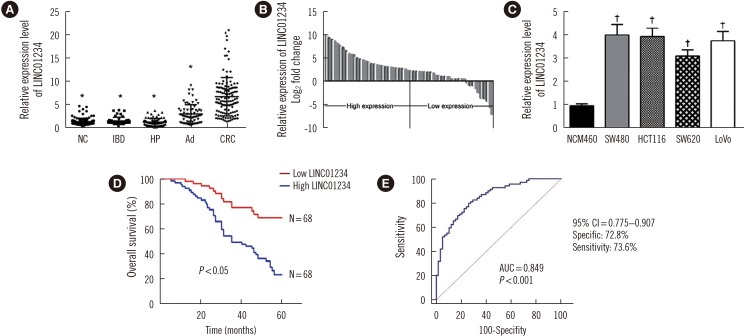
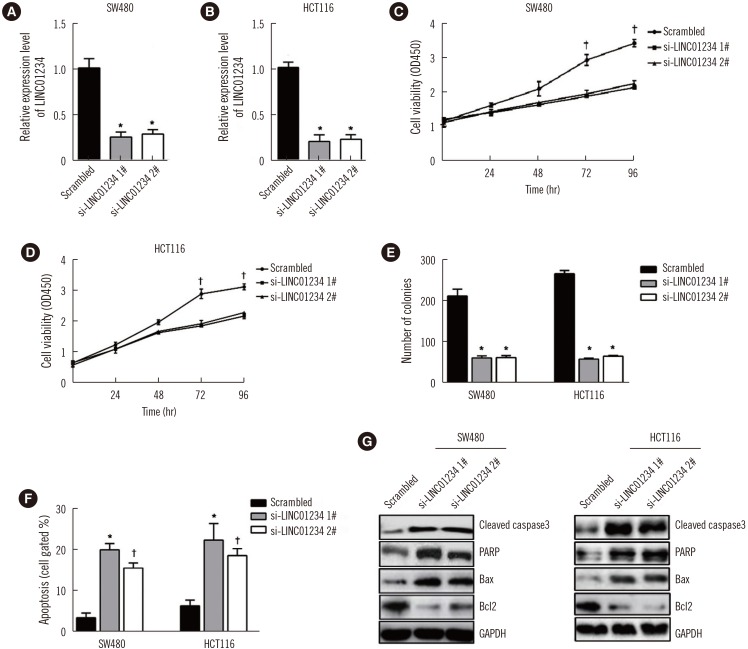
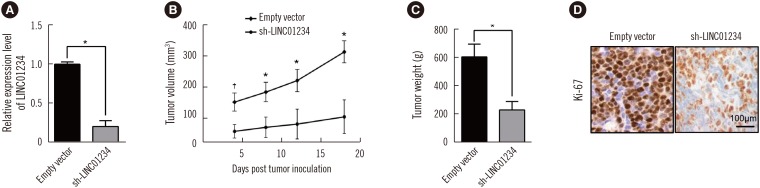
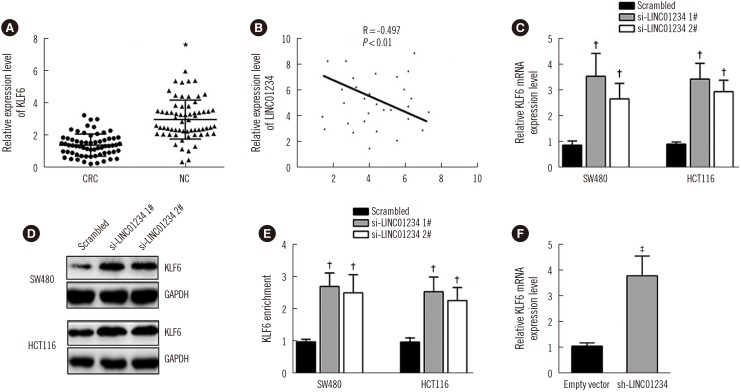
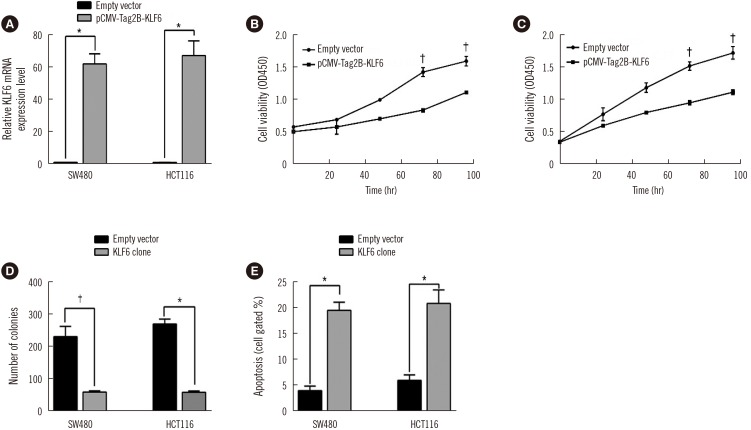
LINC01234 expression was significantly upregulated in CRC tissues and CRC cell lines than in non-tumor tissues and normal epithelial cells, respectively. LINC01234 was associated with high tumor stage, larger tumor size, and metastasis. Patients with higher LINC01234 expression showed reduced OS. Cell proliferation was inhibited by LINC01234 knockdown, whereas apoptosis was enhanced. Mice injected with SW480 cells with LINC01234 knockdown displayed decreased tumor volume, weight, and Ki-67 levels compared with those injected with control cells. KLF6 was negatively regulated by LINC01234. Overexpression of KLF6 showed effects similar to those observed following LINC01234 knockdown on cell proliferation and apoptosis.
CONCLUSIONS
LINC01234 could be a prognostic biomarker in CRC patients. Upregulation of LINC01234 in CRC promotes tumor development through negative regulation of KLF6.
MeSH Terms
-
Adenoma
Animals
Apoptosis
Blotting, Western
Cell Line
Cell Proliferation
Colorectal Neoplasms*
Epithelial Cells
Genes, Tumor Suppressor
Humans
Inflammatory Bowel Diseases
Mice
Mice, Nude
Neoplasm Metastasis
Polymerase Chain Reaction
Polyps
Prognosis*
Reverse Transcription
RNA, Long Noncoding*
RNA, Messenger
Tumor Burden
Up-Regulation
RNA, Long Noncoding
RNA, Messenger
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30. PMID: 29313949.2. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016; 25:16–27. PMID: 26667886.3. Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018; 46:D794–D801. PMID: 29126249.4. Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010; 42:1113–1117. PMID: 21057500.5. Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012; 7:e47998. PMID: 23133536.6. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. PMID: 20393566.7. Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012; 18:111–123. PMID: 22128341.8. Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013; 110:2876–2881. PMID: 23382218.9. Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014; 30:439–452. PMID: 25218058.10. Cabanski CR, White NM, Dang HX, Silva-Fisher JM, Rauck CE, Cicka D, et al. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015; 12:628–642. PMID: 25864709.11. Ghaleb AM, Yang VW. The pathobiology of Krüppel-like factors in colorectal cancer. Curr Colorectal Cancer Rep. 2008; 4:59–64. PMID: 18504508.12. He Z, Dang J, Song A, Cui X, Ma Z, Zhang Z. Identification of LINC01234 and MIR210HG as novel prognostic signature for colorectal adenocarcinoma. J Cell Physiol. 2019; 234:6769–6777. PMID: 30362555.13. Yang F, Liu YH, Dong SY, Yao ZH, Lv L, Ma RM, et al. Co-expression networks revealed potential core lncRNAs in the triple-negative breast cancer. Gene. 2016; 591:471–477. PMID: 27380926.14. Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018; 24:2002–2014. PMID: 29386218.15. Ghaffar M, Khodahemmati S, Li J, Shahzad M, Wang M, Wang Y, et al. Long non-coding RNA LINC01234 regulates proliferation, invasion and apoptosis in esophageal cancer cells. J Cancer. 2018; 9:4242–4249. PMID: 30519325.16. Cho YG, Choi BJ, Song JW, Kim SY, Nam SW, Lee SH, et al. Aberrant expression of Krüppel-like factor 6 protein in colorectal cancers. World J Gastroenterol. 2006; 12:2250–2253. PMID: 16610031.17. Zhang Q, Tan XP, Yuan YS, Hu CM, He CH, Wang WZ, et al. Decreased expression of KLF6 and its significance in gastric carcinoma. Med Oncol. 2010; 27:1295–1302. PMID: 19967571.18. DiFeo A, Feld L, Rodriguez E, Wang C, Beer DG, Martignetti JA, et al. A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 2008; 68:965–970. PMID: 18250346.19. Mukai S, Hiyama T, Tanaka S, Yoshihara M, Arihiro K, Chayama K. Involvement of Krüppel-like factor 6 (KLF6) mutation in the development of nonpolypoid colorectal carcinoma. World J Gastroenterol. 2007; 13:3932–3938. PMID: 17663506.20. Liu JX, Li W, Li JT, Liu F, Zhou L. Screening key long non-coding RNAs in early-stage colon adenocarcinoma by RNA-sequencing. Epigenomics. 2018; 10:1215–1228. PMID: 30182733.21. Xu Y, Jiang X, Cui Y. Upregulated long noncoding RNA PANDAR predicts an unfavorable prognosis and promotes tumorigenesis in cholangiocarcinoma. Onco Targets Ther. 2017; 10:2873–2883. PMID: 28652769.22. Prigge ES, Toth C, Dyckhoff G, Wagner S, Müller F, Wittekindt C, et al. p16(INK4a) /Ki-67 co-expression specifically identifies transformed cells in the head and neck region. Int J Cancer. 2015; 136:1589–1599. PMID: 25104331.23. Cho YG, Kim CJ, Park CH, Yang YM, Kim SY, Nam SW, et al. Genetic alterations of the KLF6 gene in gastric cancer. Oncogene. 2005; 24:4588–4590. PMID: 15824733.24. Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001; 294:2563–2566. PMID: 11752579.25. DiFeo A, Narla G, Hirshfeld J, Camacho-Vanegas O, Narla J, Rose SL, et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin Cancer Res. 2006; 12:3730–3739. PMID: 16778100.26. Hatami R, Sieuwerts AM, Izadmehr S, Yao Z, Qiao RF, Papa L, et al. KLF6-SV1 drives breast cancer metastasis and is associated with poor survival. Sci Transl Med. 2013; 5:169ra12.27. Narla G, DiFeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005; 65:1213–1222. PMID: 15735005.28. Reeves HL, Narla G, Ogunbiyi O, Haq AI, Katz A, Benzeno S, et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology. 2004; 126:1090–1103. PMID: 15057748.29. Zhang B, Guo DD, Zheng JY, Wu YA. Expression of KLF6-SV2 in colorectal cancer and its impact on proliferation and apoptosis. Eur J Cancer Prev. 2018; 27:20–26. PMID: 29084019.30. Cho YG, Choi BJ, Kim CJ, Song JW, Kim SY, Nam SW, et al. Genetic alterations of the KLF6 gene in colorectal cancers. APMIS. 2006; 114:458–464. PMID: 16856969.31. Benzeno S, Narla G, Allina J, Cheng GZ, Reeves HL, Banck MS, et al. Cyclin-dependent kinase inhibition by the KLF6 tumor suppressor protein through interaction with cyclin D1. Cancer Res. 2004; 64:3885–3891. PMID: 15172998.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- LINC00703 Acts as a Tumor Suppressor via Regulating miR-181a/KLF6 Axis in Gastric Cancer
- Regulation Mechanism of Long Noncoding RNAs in Colon Cancer Development and Progression
- Long Non-Coding RNA LINC00525 Promotes the Stemness and Chemoresistance of Colorectal Cancer by Targeting miR-507/ELK3 Axis
- c-Myc-Induced Long Non-Coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer
- What Is the Significance of Long Non-coding RNA HOX Transcript Antisense Intergenic RNA in Gastric Cancer?