J Breast Cancer.
2019 Dec;22(4):533-547. 10.4048/jbc.2019.22.e54.
c-Myc-Induced Long Non-Coding RNA Small Nucleolar RNA Host Gene 7 Regulates Glycolysis in Breast Cancer
- Affiliations
-
- 1Department of General Surgery, the Second Hospital of Hebei Medical University, Shijiazhuang, China. gh126123@126.com
- KMID: 2470891
- DOI: http://doi.org/10.4048/jbc.2019.22.e54
Abstract
- PURPOSE
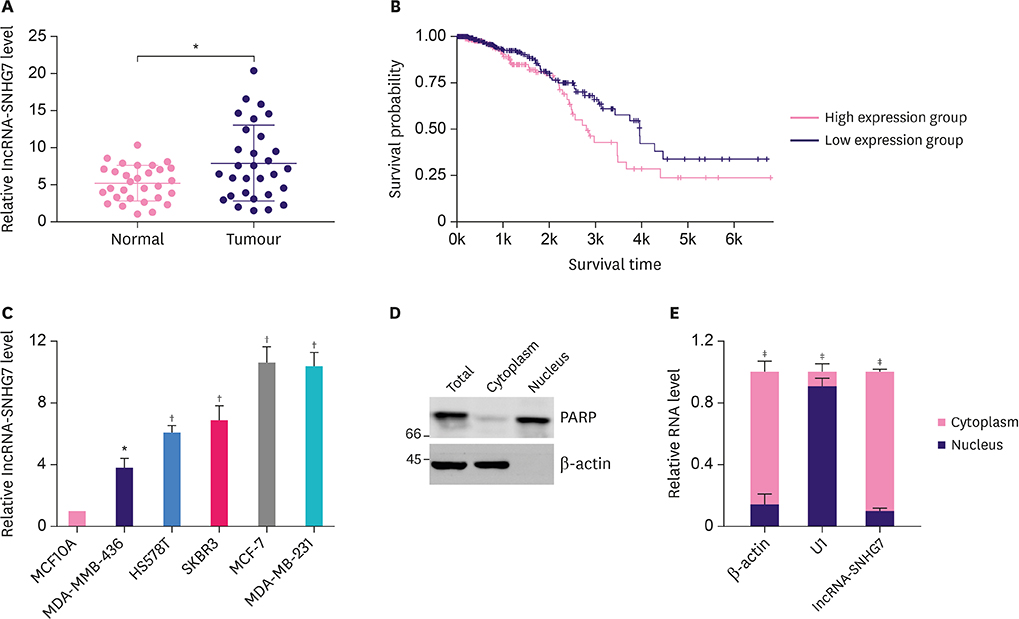
Recent studies have shown that long non-coding RNA (lncRNA) play an important role in cancer metabolism and development. The lncRNA small nucleolar RNA host gene 7 (SNHG7) was reported to be upregulated in colorectal cancer and contribute to its progression. In the current study, we investigated the role of lncRNA-SNHG7 in breast cancer and explored the underlying mechanism.
METHODS
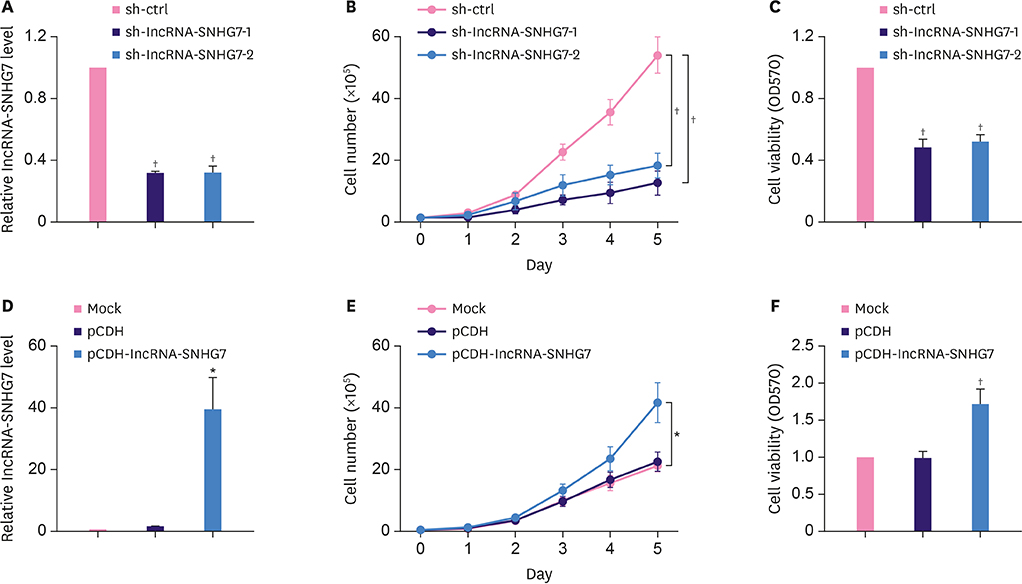
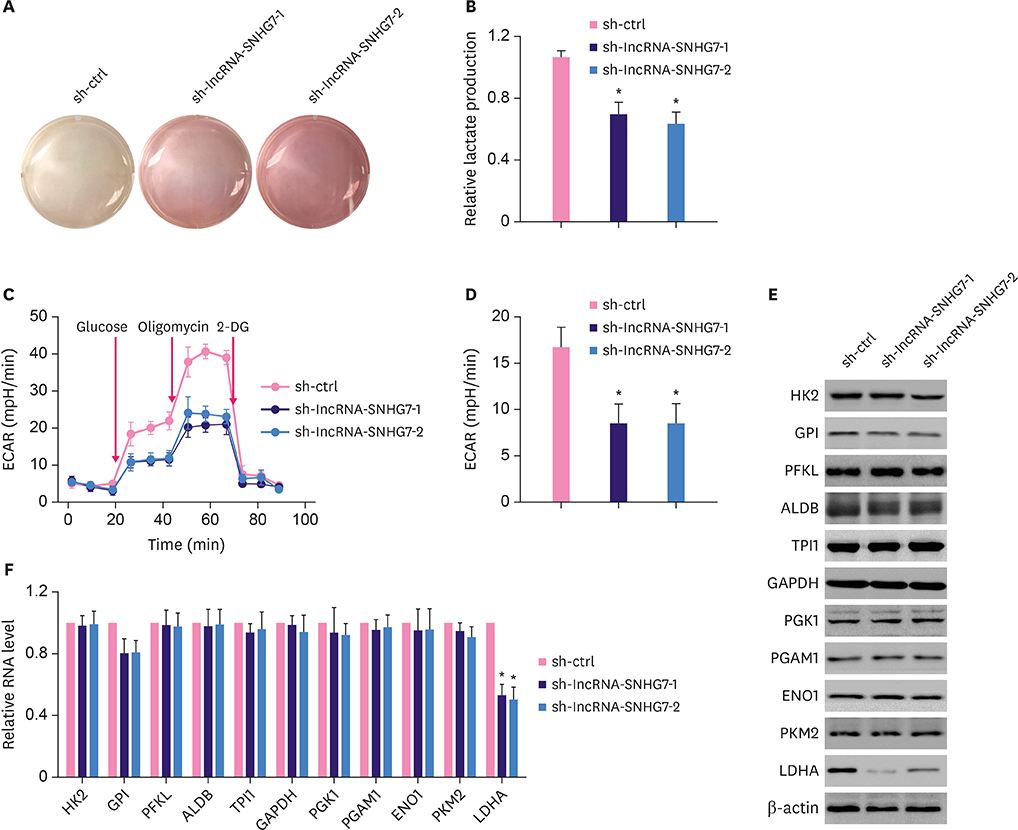
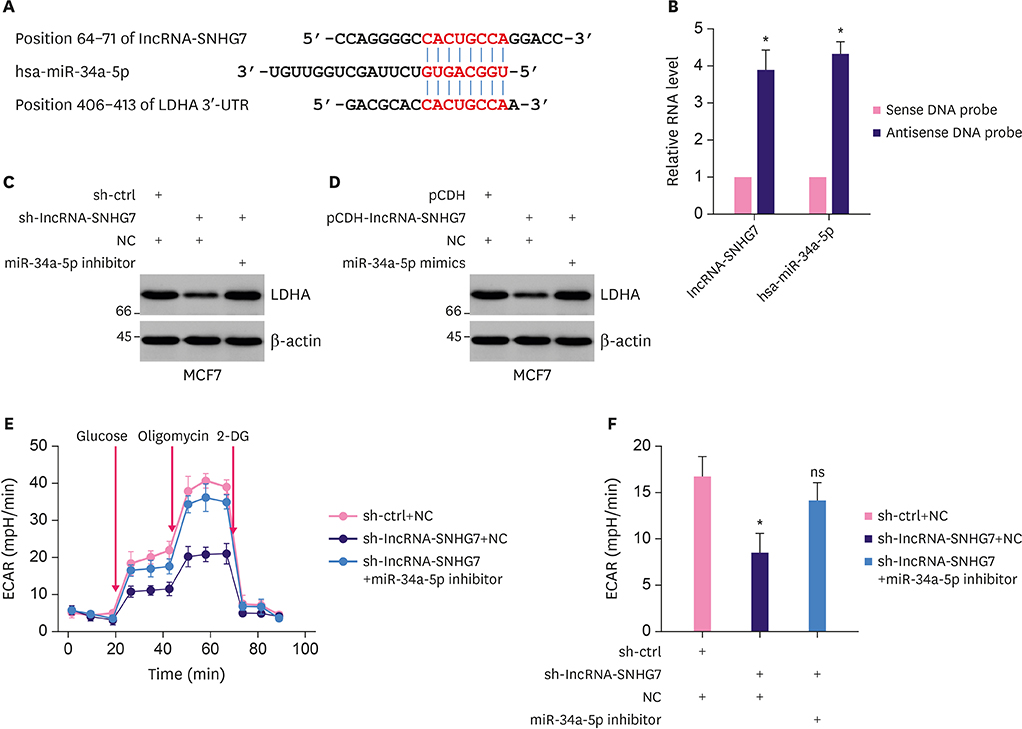
We monitored the expression of lncRNA-SNHG7 in breast cancer tissues and breast cancer cell lines. We evaluated the effects of lncRNA-SNHG7 on cell proliferation and glycolysis in breast cancer cells by knocking down or overexpressing lncRNA-SNHG7. We searched for the potential microRNA (miRNA) target of lncRNA-SNHG7 and evaluated the effects of the target miRNA on glycolysis. We evaluated the potential regulation of lncRNA-SNHG7 by c-Myc.
RESULTS
LncRNA-SNHG7 was up-regulated in both breast cancer tissues and breast cancer cell lines. Knocking down lncRNA-SNHG7 inhibited breast cancer cell proliferation while overexpressing lncRNA-SNHG7 enhanced cell proliferation. Knocking down lncRNA-SNHG7 resulted in decreased expression of lactate dehydrogenase A (LDHA) and decreased glycolysis. LncRNA-SNHG7 targeted miR-34a-5p to regulate LDHA expression and glycolysis. c-Myc bound to promoter of lncRNA-SNHG7 and positively regulated lncRNA-SNHG7 expression.
CONCLUSION
We demonstrated that c-Myc regulated glycolysis through the lncRNA-SNHG7/miR-34a-5p/LDHA axis in breast cancer cells.
Keyword
MeSH Terms
Figure
Reference
-
1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014; 64:9–29.
Article2. Coughlin SS, Ekwueme DU. Breast cancer as a global health concern. Cancer Epidemiol. 2009; 33:315–318.
Article3. Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol. 2014; 15:e279–e289.
Article4. Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013; 12:152.
Article5. Dang CV, Hamaker M, Sun P, Le A, Gao P. Therapeutic targeting of cancer cell metabolism. J Mol Med (Berl). 2011; 89:205–212.
Article6. Birsoy K, Sabatini DM, Possemato R. Untuning the tumor metabolic machine: Targeting cancer metabolism: a bedside lesson. Nat Med. 2012; 18:1022–1023.
Article7. Hamilton MJ, Young MD, Sauer S, Martinez E. The interplay of long non-coding RNAs and MYC in cancer. AIMS Biophys. 2015; 2:794–809.
Article8. Xing Z, Zhang Y, Liang K, Yan L, Xiang Y, Li C, et al. Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Res. 2018; 78:4524–4532.
Article9. Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. C-Myc and cancer metabolism. Clin Cancer Res. 2012; 18:5546–5553.
Article10. Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011; 3:83–92.11. Singh PK, Mehla K, Hollingsworth MA, Johnson KR. Regulation of aerobic glycolysis by microRNAs in cancer. Mol Cell Pharmacol. 2011; 3:125–134.12. Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol Cell. 2014; 53:88–100.
Article13. Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015; 75:3728–3737.
Article14. Li Y, Zeng C, Hu J, Pan Y, Shan Y, Liu B, et al. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J Hematol Oncol. 2018; 11:89.15. Herter EK, Stauch M, Gallant M, Wolf E, Raabe T, Gallant P. SnoRNAs are a novel class of biologically relevant Myc targets. BMC Biol. 2015; 13:25.
Article16. She K, Huang J, Zhou H, Huang T, Chen G, He J. LncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016; 36:2673–2680.
Article17. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009; 10:155–159.
Article18. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Methods Mol Biol. 2016; 1402:271–286.
Article19. Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010; 17:193–199.
Article20. Kato M, Paranjape T, Müller RU, Nallur S, Gillespie E, Keane K, et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009; 28:2419–2424.
Article21. Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, et al. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010; 31:1037–1044.
Article22. Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH, et al. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Arch Med Res. 2012; 43:514–521.
Article23. Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011; 17:211–215.
Article24. Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008; 582:1564–1568.
Article25. Yamakuchi M, Ferlito M, Lowenstein CJ. MiR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008; 105:13421–13426.
Article26. Miao P, Sheng S, Sun X, Liu J, Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 2013; 65:904–910.
Article27. Xiao X, Huang X, Ye F, Chen B, Song C, Wen J, et al. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci Rep. 2016; 6:21735.
Article28. Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008; 22:2755–2766.
Article29. He X, Tan X, Wang X, Jin H, Liu L, Ma L, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014; 35:12181–12188.
Article30. Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017; 9:533–545.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Erratum: c-Myc-Induced Long Non-Coding RNA SNHG7 Regulates Glycolysis in Breast Cancer
- Roles of Oncogenic Long Non-coding RNAs in Cancer Development
- Role of Long Non-coding Ribonucleic Acid in Gastrointestinal Cancer
- What Is the Significance of Long Non-coding RNA HOX Transcript Antisense Intergenic RNA in Gastric Cancer?
- Application of Long Non-coding RNAs in Gastric Cancer