Lab Anim Res.
2018 Dec;34(4):166-175. 10.5625/lar.2018.34.4.166.
CRISPR/Cas9-mediated knockout of Rag-2 causes systemic lymphopenia with hypoplastic lymphoid organs in FVB mice
- Affiliations
-
- 1Graduate School of Translational Medicine, Seoul National University College of Medicine, Seoul, Korea. bckang@snu.ac.kr
- 2Department of Experimental Animal Research, Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea.
- 3Biomedical Center for Animal Resource and Development, Seoul National University, College of Medicine, Seoul, Korea.
- 4Designed Animal and Transplantation Research Institute, Institute of Green Bio Science Technology, Seoul National University, Pyeongchang-gun, Gangwon-do, Korea.
- 5Department of Biotechnology, The Catholic University of Korea, Bucheon, Gyeonggi-do, Korea.
- 6College of Medicine Severance Biomedical Science Institute, Yonsei University, Seoul, Korea.
- 7Department of Biochemistry, Yonsei University, Seoul, Korea.
- KMID: 2459292
- DOI: http://doi.org/10.5625/lar.2018.34.4.166
Abstract
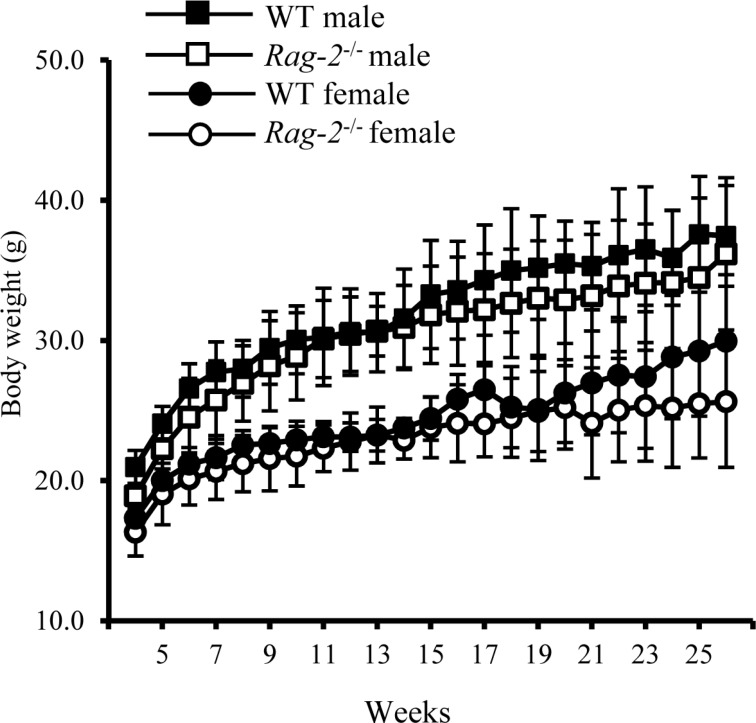
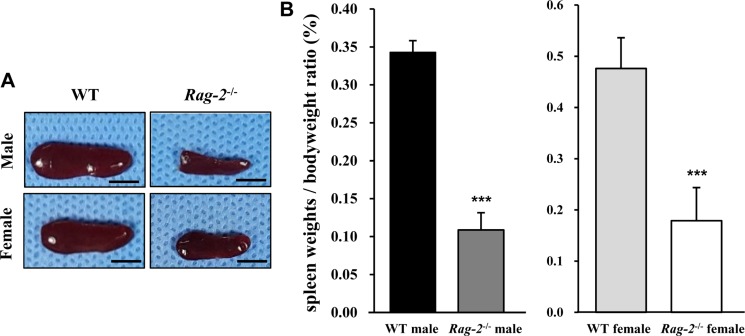
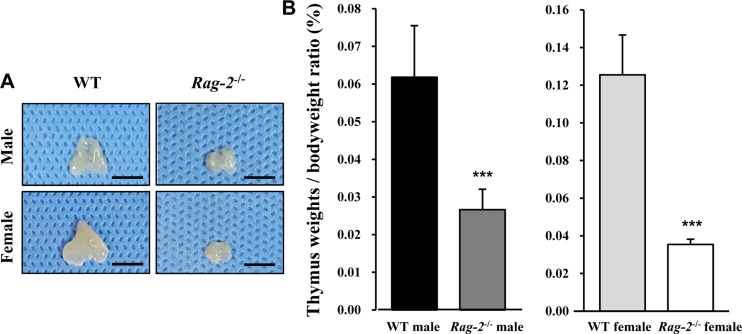
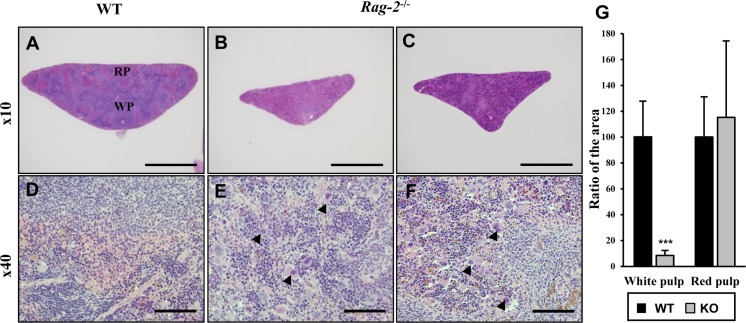
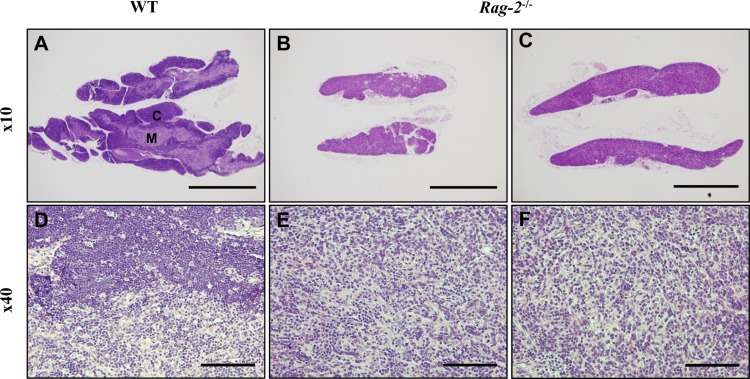
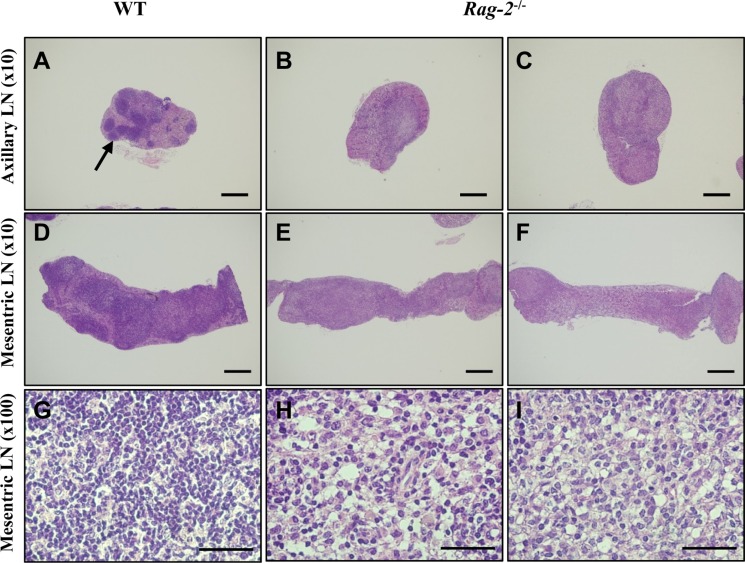
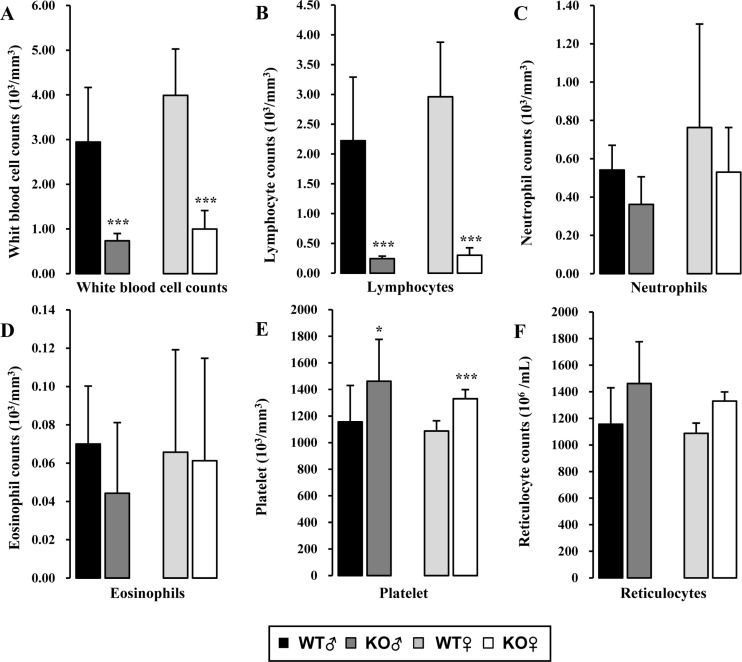
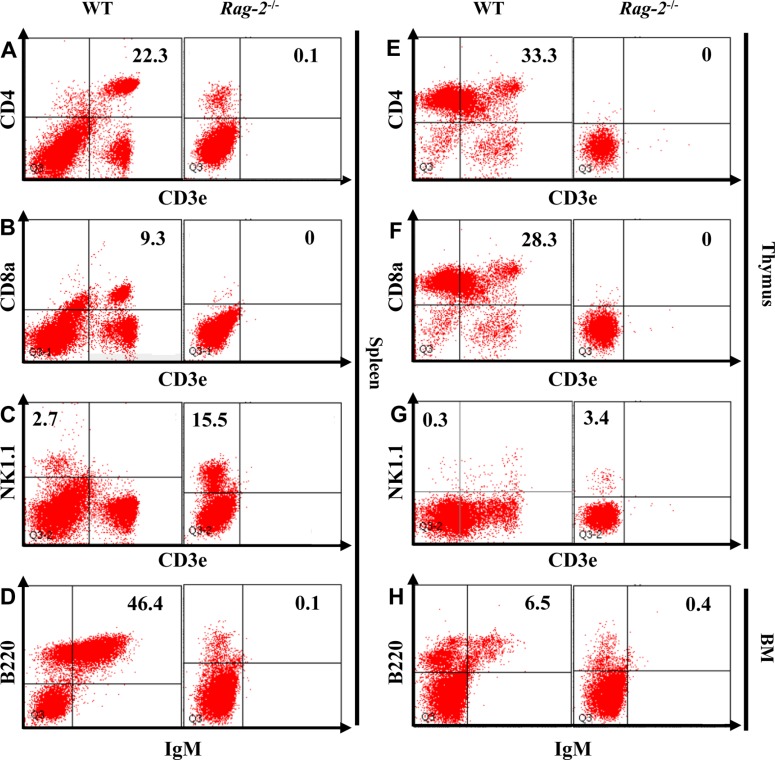
- Recombination activating gene-2 (RAG-2) plays a crucial role in the development of lymphocytes by mediating recombination of T cell receptors and immunoglobulins, and loss of RAG-2 causes severe combined immunodeficiency (SCID) in humans. RAG-2 knockout mice created using homologous recombination in ES cells have served as a valuable immunodeficient platform, but concerns have persisted on the specificity of RAG-2-related phenotypes in these animals due to the limitations associated with the genome engineering method used. To precisely investigate the function of RAG-2, we recently established a new RAG-2 knockout FVB mouse line (RAG-2(−/−)) manifesting lymphopenia by employing a CRISPR/Cas9 system at Center for Mouse Models of Human Disease. In this study, we further characterized their phenotypes focusing on histopathological analysis of lymphoid organs. RAG-2(−/−) mice showed no abnormality in development compared to their WT littermates for 26 weeks. At necropsy, gross examination revealed significantly smaller spleens and thymuses in RAG-2(−/−) mice, while histopathological investigation revealed hypoplastic white pulps with intact red pulps in the spleen, severe atrophy of the thymic cortex and disappearance of follicles in lymph nodes. However, no perceivable change was observed in the bone marrow. Moreover, our analyses showed a specific reduction of lymphocytes with a complete loss of mature T cells and B cells in the lymphoid organs, while natural killer cells and splenic megakaryocytes were increased in RAG-2(−/−) mice. These findings indicate that our RAG-2(−/−) mice show systemic lymphopenia with the relevant histopathological changes in the lymphoid organs, suggesting them as an improved Rag-2-related immunodeficient model.
MeSH Terms
-
Animals
Atrophy
B-Lymphocytes
Bone Marrow
Genome
Homologous Recombination
Humans
Immunoglobulins
Killer Cells, Natural
Lymph Nodes
Lymphocytes
Lymphopenia*
Megakaryocytes
Methods
Mice*
Mice, Knockout
Negotiating
Phenotype
Receptors, Antigen, T-Cell
Recombination, Genetic
Sensitivity and Specificity
Severe Combined Immunodeficiency
Spleen
T-Lymphocytes
Thymus Gland
Immunoglobulins
Receptors, Antigen, T-Cell
Figure
Cited by 1 articles
-
CRISPR/Cas9-mediated knockout of
CD47 causes hemolytic anemia with splenomegaly in C57BL/6 mice
Joo-Il Kim, Jin-Sung Park, Jina Kwak, Hyun-Jin Lim, Soo-Kyung Ryu, Euna Kwon, Kang-Min Han, Ki-Taek Nam, Han-Woong Lee, Byeong-Cheol Kang
Lab Anim Res. 2018;34(4):302-310. doi: 10.5625/lar.2018.34.4.302.
Reference
-
1. Nalefski EA, Kasibhatla S, Rao A. Functional analysis of the antigen binding site on the T cell receptor alpha chain. J Exp Med. 1992; 175(6):1553–1563. PMID: 1588281.
Article2. Tonegawa S. Somatic generation of antibody diversity. Nature. 1983; 302(5909):575–581. PMID: 6300689.
Article3. Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002; 71(1):101–132. PMID: 12045092.
Article4. McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995; 83(3):387–395. PMID: 8521468.
Article5. Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990; 248(4962):1517–1523. PMID: 2360047.6. Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989; 59(6):1035–1048. PMID: 2598259.7. Xu K, Liu H, Shi Z, Song G, Zhu X, Jiang Y, Zhou Z, Liu X. Disruption of the RAG2 zinc finger motif impairs protein stability and causes immunodeficiency. Eur J Immunol. 2016; 46(4):1011–1019. PMID: 26692406.
Article8. Villa A, Santagata S, Bozzi F, Imberti L, Notarangelo LD. Omenn syndrome: a disorder of Rag1 and Rag2 genes. J Clin Immunol. 1999; 19(2):87–97. PMID: 10226883.9. Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992; 68(5):855–867. PMID: 1547487.10. Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, Heike T, Nakahata T. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002; 100(9):3175–3182. PMID: 12384415.11. Skarnes WC. Is mouse embryonic stem cell technology obsolete? Genome Biol. 2015; 16(1):109. PMID: 26013980.
Article12. Valera A, Perales JC, Hatzoglou M, Bosch F. Expression of the neomycin-resistance (neo) gene induces alterations in gene expression and metabolism. Hum Gene Ther. 1994; 5(4):449–456. PMID: 7914094.
Article13. Scacheri PC, Crabtree JS, Novotny EA, Garrett-Beal L, Chen A, Edgemon KA, Marx SJ, Spiegel AM, Chandrasekharappa SC, Collins FS. Bidirectional transcriptional activity of PGK-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis. 2001; 30(4):259–263. PMID: 11536432.
Article14. Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013; 153(4):910–918. PMID: 23643243.
Article15. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339(6121):819–823. PMID: 23287718.
Article16. Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013; 31(3):233–239. PMID: 23360965.
Article17. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337(6096):816–821. PMID: 22745249.
Article18. Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154(6):1380–1389. PMID: 23992846.
Article19. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346(6213):1258096. PMID: 25430774.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- CRISPR-Cas9 system in autosomal dominant polycystic kidney disease: a comprehensive review
- Generation of knockout mouse models of cyclin-dependent kinase inhibitors by engineered nuclease-mediated genome editing
- The length of guide RNA and target DNA heteroduplex effects on CRISPR/Cas9 mediated genome editing efficiency in porcine cells
- CRISPR/Cas9-mediated generation of a Plac8 knockout mouse model
- Genome editing: the road of CRISPR/Cas9 from bench to clinic