Yonsei Med J.
2019 Jun;60(6):500-508. 10.3349/ymj.2019.60.6.500.
MiR-218-5p Suppresses the Killing Effect of Natural Killer Cell to Lung Adenocarcinoma by Targeting SHMT1
- Affiliations
-
- 1Department of Oncology, The Affiliated Renhe Hospital of China Three Gorges University, Yichang, China.
- 2Department One of Medical Oncology, Jing Men No.2 People's Hospital, Jing Men, China. nanazhai886@163.com
- 3Internal Medicine, Changyang Tujia Autonomous District People's Hospital, Yichang, China.
- KMID: 2446954
- DOI: http://doi.org/10.3349/ymj.2019.60.6.500
Abstract
- PURPOSE
Lung adenocarcinoma (LA) is one of the major types of lung cancer. MicroRNAs (miRNAs) play an essential role in regulating responses of natural killer (NK) cells to cancer malignancy. However, the mechanism of miR-218-5p involved in the killing effect of NK cells to LA cells remains poorly understood.
MATERIALS AND METHODS
The expression of miR-218-5p was examined by quantitative real-time polymerase chain reaction (qRT-PCR). Serine hydroxymethyl transferase 1 (SHMT1) level was detected by qRT-PCR or western blots. Cytokines production of interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) were detected by ELISA. The killing effect of NK cells to LA cells was investigated using lactate dehydrogenase cytotoxicity assay kit. The interaction of miR-218-5p and SHMT1 was probed by luciferase activity assay. Xenograft model was established to investigate the killing effect of NK cells in vivo.
RESULTS
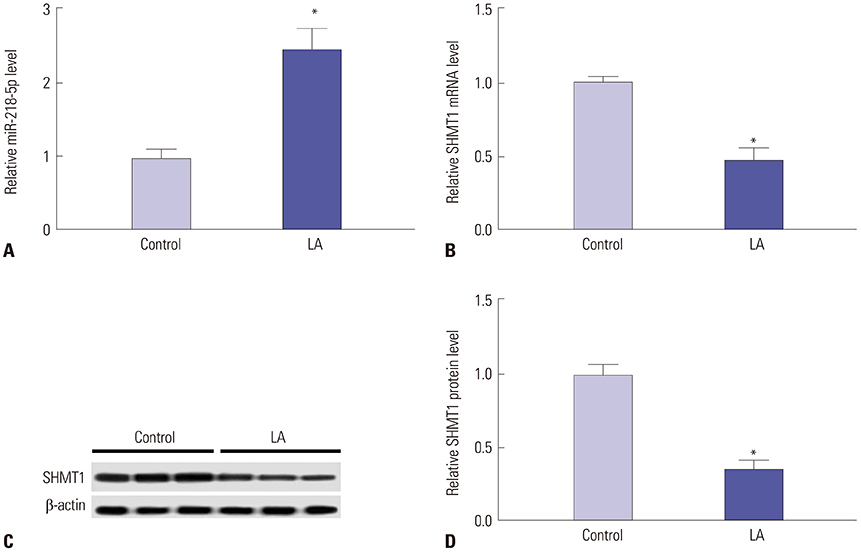
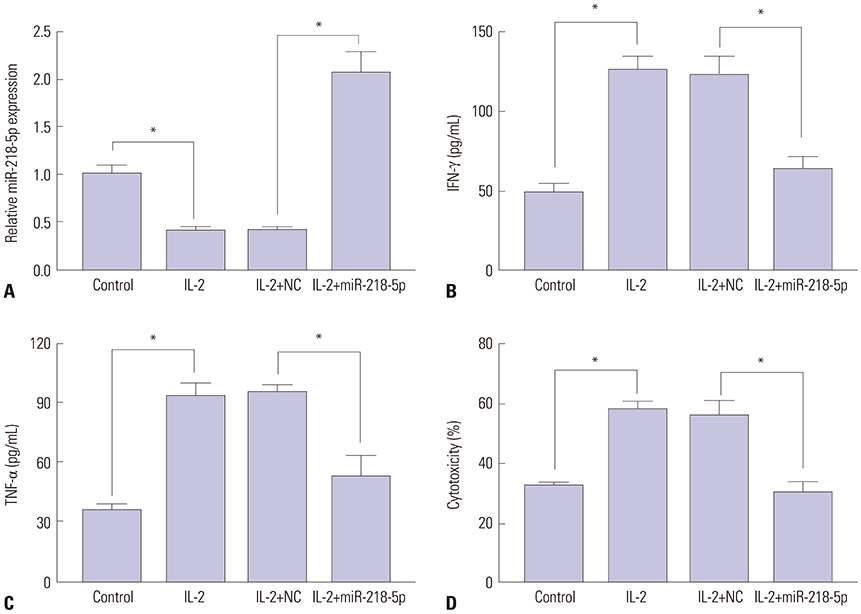
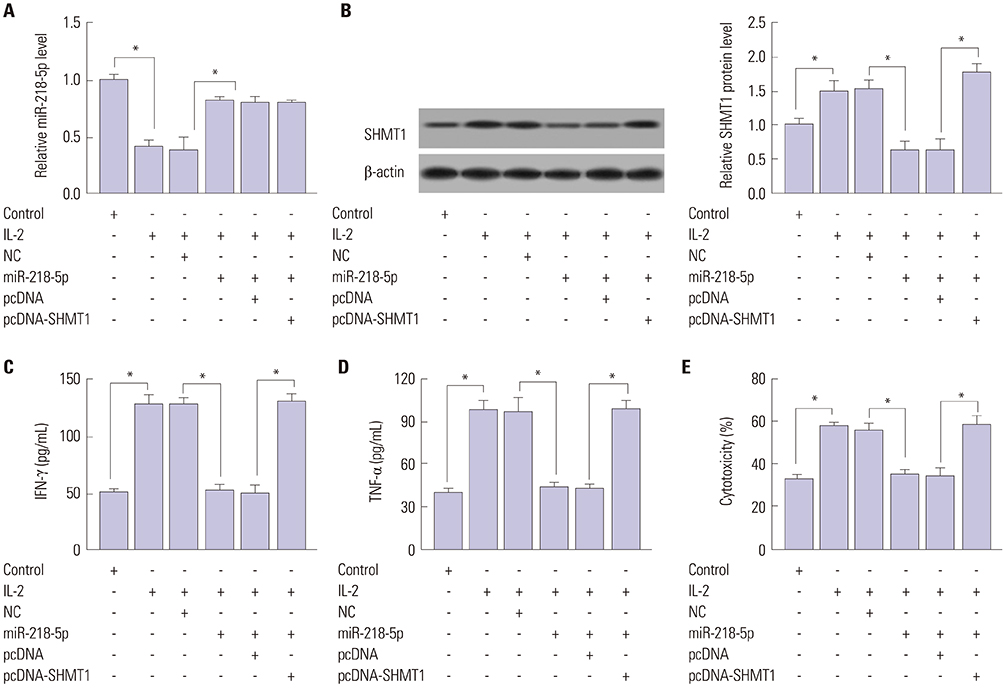
miR-218-5p was enhanced and SHMT1 was inhibited in NK cells of LA patients, whereas stimulation of interleukin-2 (IL-2) reversed their abundances. Addition of miR-218-5p reduced IL-2-induced cytokines expression and cytotoxicity in NK-92 against LA cells. Moreover, SHMT1 was negatively regulated by miR-218-5p and attenuated miR-218-5p-mediated effect on cytotoxicity, IFN-γ and TNF-α secretion in IL-2-activated NK cells. In addition, miR-218-5p exhaustion inhibited tumor growth by promoting killing effect of NK cells.
CONCLUSION
miR-218-5p suppresses the killing effect of NK cells to LA cells by targeting SHMT1, providing a potential target for LA treatment by ameliorating NK cells function.
Keyword
MeSH Terms
-
Adenocarcinoma*
Blotting, Western
Cytokines
Enzyme-Linked Immunosorbent Assay
Heterografts
Homicide*
Humans
Interleukin-2
Killer Cells, Natural*
L-Lactate Dehydrogenase
Luciferases
Lung Neoplasms
Lung*
MicroRNAs
Necrosis
Real-Time Polymerase Chain Reaction
Serine
Transferases
Cytokines
Interleukin-2
L-Lactate Dehydrogenase
Luciferases
MicroRNAs
Serine
Transferases
Figure
Reference
-
1. Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017; 3:524–548.2. Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int J Mol Sci. 2018; 19:1259.
Article3. Donington JS, Kim YT, Tong B, Moreira AL, Bessich J, Weiss KD, et al. Progress in the management of early-stage non-small cell lung cancer in 2017. J Thorac Oncol. 2018; 13:767–778.
Article4. Shroff GS, de Groot PM, Papadimitrakopoulou VA, Truong MT, Carter BW. Targeted therapy and immunotherapy in the treatment of non-small cell lung cancer. Radiol Clin North Am. 2018; 56:485–495.
Article5. Aktas¸ ON, Öztürk AB, Erman B, Erus S, Tanju S, Dilege Ş. Role of natural killer cells in lung cancer. J Cancer Res Clin Oncol. 2018; 144:997–1003.
Article6. Le Maux Chansac B, Missé D, Richon C, Vergnon I, Kubin M, Soria JC, et al. Potentiation of NK cell-mediated cytotoxicity in human lung adenocarcinoma: role of NKG2D-dependent pathway. Int Immunol. 2008; 20:801–810.
Article7. Wang R, Jaw JJ, Stutzman NC, Zou Z, Sun PD. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J Leukoc Biol. 2012; 91:299–309.
Article8. Maemura K, Watanabe K, Ando T, Hiyama N, Sakatani T, Amano Y, et al. Altered editing level of microRNAs is a potential biomarker in lung adenocarcinoma. Cancer Sci. 2018; 109:3326–3335.
Article9. Shirshev SV, Nekrasova IV, Gorbunova OL, Orlova EG, Maslennikova IL. MicroRNA in hormonal mechanisms of regulation of NK cell function. Dokl Biochem Biophys. 2017; 474:168–172.
Article10. Rizzo R, Soffritti I, D'Accolti M, Bortolotti D, Di Luca D, Caselli E. HHV-6A/6B infection of NK cells modulates the expression of miRNAs and transcription factors potentially associated to impaired NK activity. Front Microbiol. 2017; 8:2143.
Article11. Donatelli SS, Zhou JM, Gilvary DL, Eksioglu EA, Chen X, Cress WD, et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014; 111:4203–4208.
Article12. Li YJ, Zhang W, Xia H, Zhang BS, Chen P, Zhao YL, et al. miR-218 suppresses epithelial-to-mesenchymal transition by targeting Robo1 and Ecop in lung adenocarcinoma cells. Future Oncol. 2017; 13:2571–2582.
Article13. Victor AR, Weigel C, Scoville SD, Chan WK, Chatman K, Nemer MM, et al. Epigenetic and posttranscriptional regulation of CD16 expression during human NK cell development. J Immunol. 2018; 200:565–572.
Article14. Gupta R, Yang Q, Dogra SK, Wajapeyee N. Serine hydroxymethyl transferase 1 stimulates pro-oncogenic cytokine expression through sialic acid to promote ovarian cancer tumor growth and progression. Oncogene. 2017; 36:4014–4024.
Article15. Bahari G, Hashemi M, Naderi M, Sadeghi-Bojd S, Taheri M. Association of SHMT1 gene polymorphisms with the risk of childhood acute lymphoblastic leukemia in a sample of Iranian population. Cell Mol Biol (Noisy-le-grand). 2016; 62:45–51.16. Wang L, Lu J, An J, Shi Q, Spitz MR, Wei Q. Polymorphisms of cytosolic serine hydroxymethyltransferase and risk of lung cancer: a case-control analysis. Lung Cancer. 2007; 57:143–151.
Article17. Wu S, Zhang G, Li P, Chen S, Zhang F, Li J, et al. miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol. 2016; 37:5193–5202.
Article18. Xie H, Zhang Q, Zhou H, Zhou J, Zhang J, Jiang Y, et al. microRNA-889 is downregulated by histone deacetylase inhibitors and confers resistance to natural killer cytotoxicity in hepatocellular carcinoma cells. Cytotechnology. 2018; 70:513–521.
Article19. Zhao H, Su W, Kang Q, Xing Z, Lin X, Wu Z. Natural killer cells inhibit oxaliplatin-resistant colorectal cancer by repressing WBSCR22 via upregulating microRNA-146b-5p. Am J Cancer Res. 2018; 8:824–834.20. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017; 169:750–765.
Article21. Jasinski-Bergner S, Mandelboim O, Seliger B. The role of microRNAs in the control of innate immune response in cancer. J Natl Cancer Inst. 2014; 106:dju257.
Article22. Xu D, Han Q, Hou Z, Zhang C, Zhang J. miR-146a negatively regulates NK cell functions via STAT1 signaling. Cell Mol Immunol. 2017; 14:712–720.
Article23. Ma Y, Gong J, Liu Y, Guo W, Jin B, Wang X, et al. MicroRNA-30c promotes natural killer cell cytotoxicity via up-regulating the expression level of NKG2D. Life Sci. 2016; 151:174–181.
Article24. Liu M, Yin K, Guo X, Feng H, Yuan M, Liu Y, et al. Diphthamide biosynthesis 1 is a novel oncogene in colorectal cancer cells and is regulated by MiR-218-5p. Cell Physiol Biochem. 2017; 44:505–514.
Article25. Lu J, Ji ML, Zhang XJ, Shi PL, Wu H, Wang C, et al. MicroRNA-218-5p as a potential target for the treatment of human osteoarthritis. Mol Ther. 2017; 25:2676–2688.
Article26. Tian F, Li R, Chen Z, Shen Y, Lu J, Xie X, et al. Differentially expressed miRNAs in tumor, adjacent, and normal tissues of lung adenocarcinoma. Biomed Res Int. 2016; 2016:1428271.
Article27. Marcais A, Viel S, Grau M, Henry T, Marvel J, Walzer T. Regulation of mouse NK cell development and function by cytokines. Front Immunol. 2013; 4:450.
Article28. Pan C, Xiang L, Pan Z, Wang X, Li J, Zhuge L, et al. MiR-544 promotes immune escape through downregulation of NCR1/NKp46 via targeting RUNX3 in liver cancer. Cancer Cell Int. 2018; 18:52.
Article29. Zhang LL, Zhang LF, Shi YB. miR-24 inhibited the killing effect of natural killer cells to colorectal cancer cells by downregulating Paxillin. Biomed Pharmacother. 2018; 101:257–263.
Article30. Rahmoon MA, Youness RA, Gomaa AI, Hamza MT, Waked I, El Tayebi HM, et al. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017; 35:76–87.
Article31. Leong JW, Wagner JA, Ireland AR, Fehniger TA. Transcriptional and post-transcriptional regulation of NK cell development and function. Clin Immunol. 2017; 177:60–69.
Article32. Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X, et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017; 16:141.
Article33. Shi ZM, Wang L, Shen H, Jiang CF, Ge X, Li DM, et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene. 2017; 36:2577–2588.
Article34. Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S, et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2017; 114:11404–11409.
Article35. Ueland PM, McCann A, Midttun Ø, Ulvik A. Inflammation, vitamin B6 and related pathways. Mol Aspects Med. 2017; 53:10–27.
Article36. Gupta R, Yang Q, Dogra SK, Wajapeyee N. Serine hydroxymethyl transferase 1 stimulates pro-oncogenic cytokine expression through sialic acid to promote ovarian cancer tumor growth and progression. Oncogene. 2017; 36:4014–4024.
Article37. Lv Q, Hu JX, Li YJ, Xie N, Song DD, Zhao W, et al. MiR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol Ther. 2017; 18:142–151.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- miR-450a-5p Eliminates MGO-Induced Insulin Resistance via Targeting CREB
- MiR-322-5p Alleviates Cell Injury and Impairment of Cognitive Function in Vascular Dementia by Targeting TSPAN5
- MicroRNA-98-5p Inhibits Tumorigenesis of Hepatitis B Virus-Related Hepatocellular Carcinoma by Targeting NF-κB-Inducing Kinase
- Long Noncoding RNA Cytoskeleton Regulator RNA Suppresses Apoptosis in Hepatoma Cells by Modulating the miR-125a-5p/ HS1-Associated Protein X-1 Axis to Induce Caspase-9 Inactivation
- Down-regulation of microRNA-382-5p reduces neuropathic pain by targeting regulation of dual specificity phosphatase-1