Korean J Physiol Pharmacol.
2019 Mar;23(2):151-159. 10.4196/kjpp.2019.23.2.151.
Prediction of itching diagnostic marker through RNA sequencing of contact hypersensitivity and skin scratching stimulation mice models
- Affiliations
-
- 1Department of Physiology, Chung-Ang University College of Medicine, Seoul 06974, Korea. haena@cau.ac.kr, akdongyi01@cau.ac.kr
- 2Department of Physiology and Cell Biology, University of Nevada, Reno School of Medicine, Reno, NV 89557, USA.
- 3Department of Medicine, Chung-Ang University College of Medicine, Seoul 06974, Korea.
- KMID: 2438089
- DOI: http://doi.org/10.4196/kjpp.2019.23.2.151
Abstract
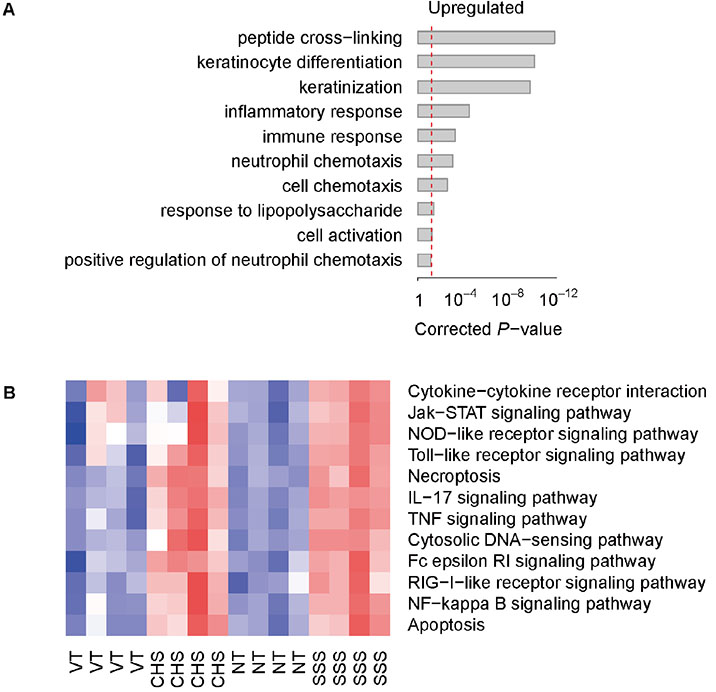
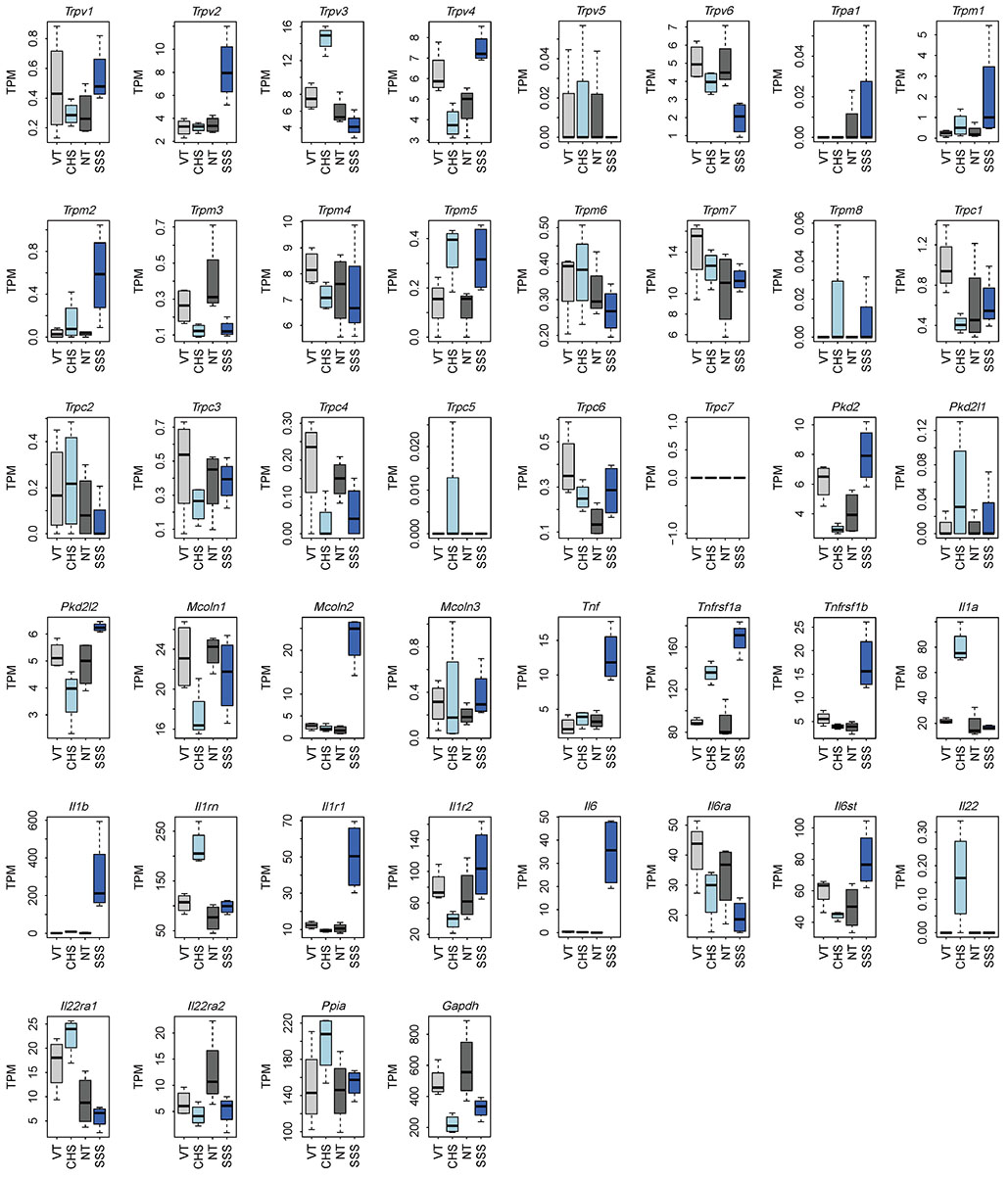
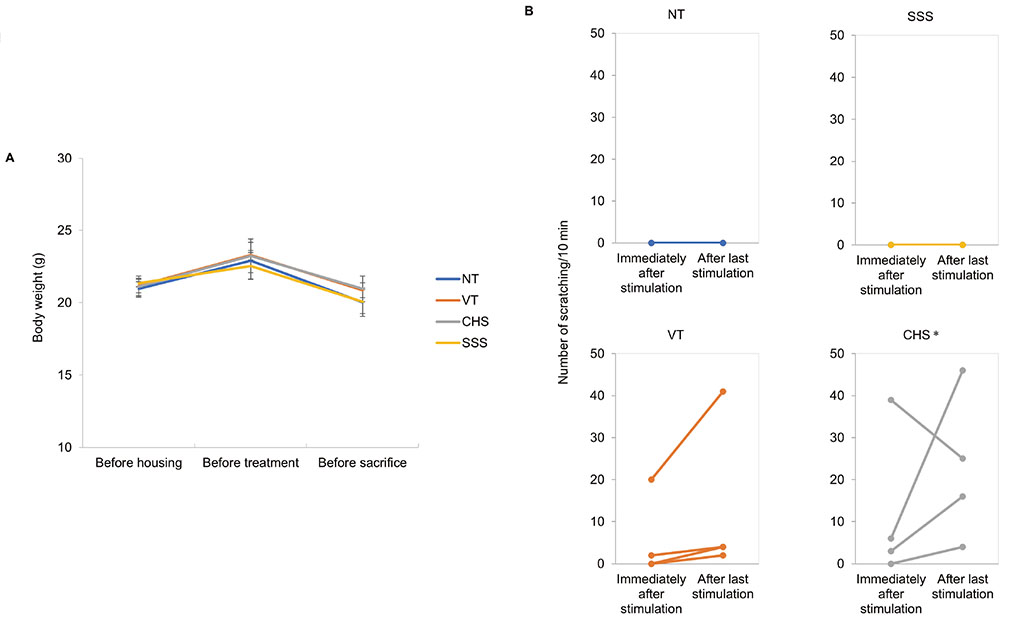
- Pruritus (itching) is classically defined as an unpleasant cutaneous sensation that leads to scratching behavior. Although the scientific criteria of classification for pruritic diseases are not clear, it can be divided as acute or chronic by duration of symptoms. In this study, we investigated whether skin injury caused by chemical (contact hypersensitivity, CHS) or physical (skin-scratching stimulation, SSS) stimuli causes initial pruritus and analyzed gene expression profiles systemically to determine how changes in skin gene expression in the affected area are related to itching. In both CHS and SSS, we ranked the Gene Ontology Biological Process terms that are generally associated with changes. The factors associated with upregulation were keratinization, inflammatory response and neutrophil chemotaxis. The Kyoto Encyclopedia of Genes and Genomes pathway shows the difference of immune system, cell growth and death, signaling molecules and interactions, and signal transduction pathways. Il1a , Il1b and Il22 were upregulated in the CHS, and Tnf, Tnfrsf1b, Il1b, Il1r1 and Il6 were upregulated in the SSS. Trpc1 channel genes were observed in representative itching-related candidate genes. By comparing and analyzing RNA-sequencing data obtained from the skin tissue of each animal model in these characteristic stages, it is possible to find useful diagnostic markers for the treatment of itching, to diagnose itching causes and to apply customized treatment.
Keyword
MeSH Terms
-
Animals
Biological Processes
Chemotaxis
Classification
Cytokines
Dermatitis, Contact*
Gene Expression
Gene Ontology
Genome
Hypersensitivity
Immune System
Interleukin-6
Mice*
Models, Animal
Neutrophils
Pruritus*
RNA*
Sensation
Sequence Analysis, RNA*
Signal Transduction
Skin*
Transcriptome
Transient Receptor Potential Channels
Up-Regulation
Wound Healing
Cytokines
Interleukin-6
RNA
Transient Receptor Potential Channels
Figure
Reference
-
1. Ständer S, Weisshaar E, Mettang T, Szepietowski JC, Carstens E, Ikoma A, Bergasa NV, Gieler U, Misery L, Wallengren J, Darsow U, Streit M, Metze D, Luger TA, Greaves MW, Schmelz M, Yosipovitch G, Bernhard JD. Clinical classification of itch: a position paper of the International Forum for the Study of Itch. Acta Derm Venereol. 2007; 87:291–294.
Article2. Moore C, Gupta R, Jordt SE, Chen Y, Liedtke WB. Regulation of pain and itch by TRP channels. Neurosci Bull. 2018; 34:120–142.
Article3. Anderson SE, Siegel PD, Meade BJ. The LLNA: a brief review of recent advances and limitations. J Allergy (Cairo). 2011; 2011:424203.
Article4. Honda T, Egawa G, Grabbe S, Kabashima K. Update of immune events in the murine contact hypersensitivity model: toward the understanding of allergic contact dermatitis. J Invest Dermatol. 2013; 133:303–315.
Article5. Yamaoka J, Kawana S. A transient unresponsive state of self-scratching behaviour is induced in mice by skin-scratching stimulation. Exp Dermatol. 2007; 16:737–745.
Article6. Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA. 2017; 8:e1364.
Article7. Zhang X. Targeting TRP ion channels for itch relief. Naunyn Schmiedebergs Arch Pharmacol. 2015; 388:389–399.
Article8. Shim WS, Tak MH, Lee MH, Kim M, Kim M, Koo JY, Lee CH, Kim M, Oh U. TRPV1 mediates histamine-induced itching via the activation of phospholipase A2 and 12-lipoxygenase. J Neurosci. 2007; 27:2331–2337.9. DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012; 28:1530–1532.
Article10. Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, et al. Ensembl 2015. Nucleic Acids Res. 2015; 43:D662–D669.
Article11. Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014; 32:462–464.
Article12. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140.
Article13. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004; 32:D258–D261.14. Yang X, Regan K, Huang Y, Zhang Q, Li J, Seiwert TY, Cohen EE, Xing HR, Lussier YA. Single sample expression-anchored mechanisms predict survival in head and neck cancer. PLoS Comput Biol. 2012; 8:e1002350.
Article15. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452.
Article16. Schmelz M. Itch and pain. Neurosci Biobehav Rev. 2010; 34:171–176.
Article17. Twycross R, Greaves MW, Handwerker H, Jones EA, Libretto SE, Szepietowski JC, Zylicz Z. Itch: scratching more than the surface. QJM. 2003; 96:7–26.
Article18. Rittié L. Cellular mechanisms of skin repair in humans and other mammals. J Cell Commun Signal. 2016; 10:103–120.
Article19. Jensen LE. Targeting the IL-1 family members in skin inflammation. Curr Opin Investig Drugs. 2010; 11:1211–1220.20. Wang XP, Schunck M, Kallen KJ, Neumann C, Trautwein C, Rose-John S, Proksch E. The interleukin-6 cytokine system regulates epidermal permeability barrier homeostasis. J Invest Dermatol. 2004; 123:124–131.
Article21. McGee HM, Schmidt BA, Booth CJ, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA, Horsley V. IL-22 promotes fibroblast-mediated wound repair in the skin. J Invest Dermatol. 2013; 133:1321–1329.
Article22. Feng J, Yang P, Mack MR, Dryn D, Luo J, Gong X, Liu S, Oetjen LK, Zholos AV, Mei Z, Yin S, Kim BS, Hu H. Sensory TRP channels contribute differentially to skin inflammation and persistent itch. Nat Commun. 2017; 8:980.
Article23. Wilson SR, Thé L, Batia LM, Beattie K, Katibah GE, McClain SP, Pellegrino M, Estandian DM, Bautista DM. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013; 155:285–295.
Article24. Belkacemi T, Niermann A, Hofmann L, Wissenbach U, Birnbaumer L, Leidinger P, Backes C, Meese E, Keller A, Bai X, Scheller A, Kirchhoff F, Philipp SE, Weissgerber P, Flockerzi V, Beck A. TRPC1- and TRPC3-dependent Ca2+ signaling in mouse cortical astrocytes affects injury-evoked astrogliosis in vivo. Glia. 2017; 65:1535–1549.25. Jing PB, Cao DL, Li SS, Zhu M, Bai XQ, Wu XB, Gao YJ. Chemokine receptor CXCR3 in the spinal cord contributes to chronic itch in mice. Neurosci Bull. 2018; 34:54–63.
Article26. Sun L, Hua Y, Vergarajauregui S, Diab HI, Puertollano R. Novel role of TRPML2 in the regulation of the innate immune response. J Immunol. 2015; 195:4922–4932.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Alteration in Contact Hypersensitivity of Mice induced by Indomethacin Treatment
- Chronic Toxoplasmosis Modulates the Induction of Contact Hypersensitivity by TNCB in Mouse Model
- Evaluation of Three Different Methods to Establish Animal Models of Acanthamoeba Keratitis
- Immunologic Mechanism of Experimental and Therapeutic Ultraviolet B Responses
- The Effect of Topical Tacrolimus in the Murine Contact Hypersensitivity and Dermatitis of Repeated Applications Induced by Diphenylcyclopropenone