Intest Res.
2018 Jul;16(3):338-345. 10.5217/ir.2018.16.3.338.
Intestinal microbiota, chronic inflammation, and colorectal cancer
- Affiliations
-
- 1Department of Internal Medicine, Hanyang University Guri Hospital, Hanyang University College of Medicine, Guri, Korea. hands@hanyang.ac.kr
- KMID: 2417645
- DOI: http://doi.org/10.5217/ir.2018.16.3.338
Abstract
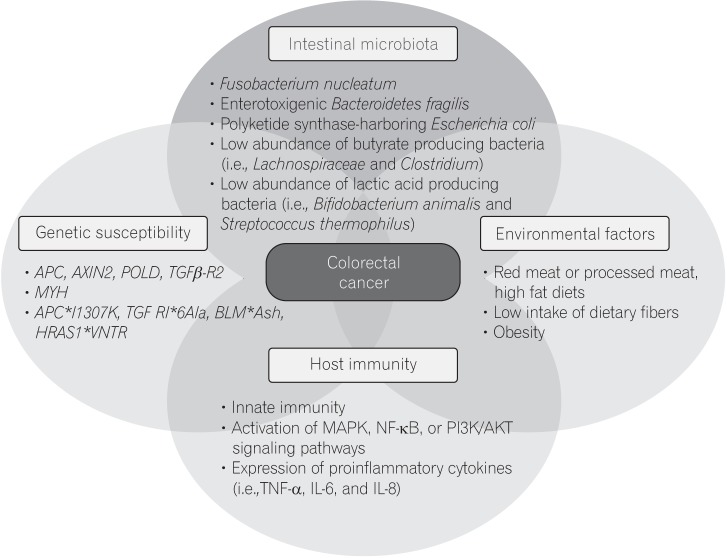
- In addition to genetic and epigenetic factors, various environmental factors, including diet, play important roles in the development of colorectal cancer (CRC). Recently, there is increasing interest in the intestinal microbiota as an environmental risk factor for CRC, because diet also influences the composition of the intestinal microbiota. The human intestinal microbiota comprises about 100 trillion microbes. This microbiome thrives on undigested dietary residues in the intestinal lumen and produces various metabolites. It is well known that the dietary risk factors for CRC are mediated by dysbiosis of the intestinal microbiota and their metabolites. In this review, we describe the bacterial taxa associated with CRC, including Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, Escherichia coli, and butyrate-producing bacteria. We also discuss the host-diet interaction in colorectal carcinogenesis.
Keyword
MeSH Terms
Figure
Cited by 2 articles
-
The role of microbiome in colorectal carcinogenesis and its clinical potential as a target for cancer treatment
Sang Hoon Kim, Yun Jeong Lim
Intest Res. 2022;20(1):31-42. doi: 10.5217/ir.2021.00034.식이가 대장암의 진행 및 예방에 미치는 영향: 영양소부터 종양 발생까지
Sang Hoon Kim, Dong Hwan Park, Yun Jeong Lim
Korean J Gastroenterol. 2023;82(2):73-83. doi: 10.4166/kjg.2023.079.
Reference
-
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. PMID: 25651787.
Article2. Perdue DG, Haverkamp D, Perkins C, Daley CM, Provost E. Geographic variation in colorectal cancer incidence and mortality, age of onset, and stage at diagnosis among American Indian and Alaska Native people, 1990-2009. Am J Public Health. 2014; 104(Suppl 3):S404–S414. PMID: 24754657.
Article3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66:7–30. PMID: 26742998.
Article4. Siegel RL, Fedewa SA, Anderson WF, et al. Colorectal cancer incidence patterns in the United States, 1974-2013. J Natl Cancer Inst. 2017; 109. DOI: 10.1093/jnci/djw322.
Article5. Ng SC, Wong SH. Colorectal cancer screening in Asia. Br Med Bull. 2013; 105:29–42. PMID: 23299409.
Article6. Shin A, Kim KZ, Jung KW, et al. Increasing trend of colorectal cancer incidence in Korea, 1999-2009. Cancer Res Treat. 2012; 44:219–226. PMID: 23341785.
Article7. Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011; 8:686–700. PMID: 22009203.
Article8. Hughes LA, Simons CC, van den Brandt PA, van Engeland M, Weijenberg MP. Lifestyle, diet, and colorectal cancer risk according to (epi)genetic instability: current evidence and future directions of molecular pathological epidemiology. Curr Colorectal Cancer Rep. 2017; 13:455–469. PMID: 29249914.
Article9. Aune D, Chan DS, Lau R, et al. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ. 2011; 343:d6617. DOI: 10.1136/bmj.d6617. PMID: 22074852.
Article10. Magalhães B, Peleteiro B, Lunet N. Dietary patterns and colorectal cancer: systematic review and meta-analysis. Eur J Cancer Prev. 2012; 21:15–23. PMID: 21946864.11. IARC monographs evaluate consumption of red meat and processed meat. International Agency for Research on Cancer Web site. Accessed Mar 1, 2018. https://www.iarc.fr/en/media-centre/pr/2015/pdfs/pr240_E.pdf .12. Le Marchand L, Kolonel LN. Cancer in Japanese migrants to Hawaii: interaction between genes and environment. Rev Epidemiol Sante Publique. 1992; 40:425–430. PMID: 1287741.13. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486:207–214. PMID: 22699609.14. Vipperla K, O'Keefe SJ. Diet, microbiota, and dysbiosis: a ‘recipe’ for colorectal cancer. Food Funct. 2016; 7:1731–1740. PMID: 26840037.
Article15. Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015; 6:6528. PMID: 25758642.
Article16. Flemer B, Warren RD, Barrett MP, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018; 67:1454–1463. PMID: 28988196.
Article17. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013; 14:207–215. PMID: 23954159.
Article18. Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013; 105:1907–1911. PMID: 24316595.
Article19. Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014; 10:766. PMID: 25432777.
Article20. Vogtmann E, Hua X, Zeller G, et al. Colorectal cancer and the human gut microbiome: reproducibility with whole-genome shotgun sequencing. PLoS One. 2016; 11:e0155362. DOI: 10.1371/journal.pone.0155362. PMID: 27171425.
Article21. Liang Q, Chiu J, Chen Y, et al. Fecal bacteria act as novel biomarkers for noninvasive diagnosis of colorectal cancer. Clin Cancer Res. 2017; 23:2061–2070. PMID: 27697996.
Article22. McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013; 8:e53653. DOI: 10.1371/journal.pone.0053653.PMID: 23335968.
Article23. Park CH, Han DS, Oh YH, Lee AR, Lee YR, Eun CS. Role of Fusobacteria in the serrated pathway of colorectal carcinogenesis. Sci Rep. 2016; 6:25271. PMID: 27125587.
Article24. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013; 14:195–206. PMID: 23954158.
Article25. Abed J, Emgård JE, Zamir G, et al. Fap2 Mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016; 20:215–225. PMID: 27512904.
Article26. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017; 170:548–563.e16. PMID: 28753429.
Article27. Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015; 42:344–355. PMID: 25680274.
Article28. Wong SH, Zhao L, Zhang X, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017; 153:1621–1633.e6. PMID: 28823860.
Article29. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014; 12:661–672. PMID: 25198138.
Article30. Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008; 19:587–593. PMID: 18061431.
Article31. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017; 66:70–78. PMID: 26408641.
Article32. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driverpassenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012; 10:575–582. PMID: 22728587.
Article33. Chen J, Pitmon E, Wang K. Microbiome, inflammation and colorectal cancer. Semin Immunol. 2017; 32:43–53. PMID: 28982615.
Article34. Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011; 6:e16393. DOI: 10.1371/journal.pone.0016393. PMID: 21297998.
Article35. Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012; 6:320–329. PMID: 21850056.
Article36. Zackular JP, Rogers MA, Ruffin MT 4th, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). 2014; 7:1112–1121. PMID: 25104642.
Article37. Sears CL, Islam S, Saha A, et al. Association of enterotoxigenic Bacteroides fragilis infection with inflammatory diarrhea. Clin Infect Dis. 2008; 47:797–803. PMID: 18680416.
Article38. O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016; 13:691–706. PMID: 27848961.39. Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009; 15:1016–1022. PMID: 19701202.
Article40. Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012; 338:120–123. PMID: 22903521.
Article41. Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014; 63:1932–1942. PMID: 24658599.
Article42. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010; 107:11537–11542. PMID: 20534522.
Article43. Tomkovich S, Yang Y, Winglee K, et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017; 77:2620–2632. PMID: 28416491.
Article44. Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011; 53:870–878. PMID: 21960713.
Article45. Boleij A, Muytjens CM, Bukhari SI, et al. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis. 2011; 203:1101–1109. PMID: 21451000.
Article46. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008; 27:104–119. PMID: 17973645.
Article47. Ruemmele FM, Schwartz S, Seidman EG, Dionne S, Levy E, Lentze MJ. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut. 2003; 52:94–100. PMID: 12477768.
Article48. Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013; 341:569–573. PMID: 23828891.
Article49. Smith EA, Macfarlane GT. Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol. 1998; 25:355–368.
Article50. Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996; 81:288–302. PMID: 8810056.
Article51. Jones RM, Luo L, Ardita CS, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013; 32:3017–3028. PMID: 24141879.
Article52. Okada T, Fukuda S, Hase K, et al. Microbiota-derived lactate accelerates colon epithelial cell turnover in starvation-refed mice. Nat Commun. 2013; 4:1654. PMID: 23552069.
Article53. Song M, Chan AT. Diet, gut microbiota, and colorectal cancer prevention: a review of potential mechanisms and promising targets for future research. Curr Colorectal Cancer Rep. 2017; 13:429–439. PMID: 29333111.
Article54. Flemer B, Lynch DB, Brown JM, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017; 66:633–643. PMID: 26992426.
Article55. Stewart JA, Chadwick VS, Murray A. Carriage, quantification, and predominance of methanogens and sulfate-reducing bacteria in faecal samples. Lett Appl Microbiol. 2006; 43:58–63. PMID: 16834722.
Article56. Hudson MJ, Tomkins AM, Wiggins HS, Drasar BS. Breath methane excretion and intestinal methanogenesis in children and adults in rural Nigeria. Scand J Gastroenterol. 1993; 28:993–998. PMID: 8284637.
Article57. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005; 308:1635–1638. PMID: 15831718.
Article58. Matarazzo F, Ribeiro AC, Faveri M, Taddei C, Martinez MB, Mayer MP. The domain Archaea in human mucosal surfaces. Clin Microbiol Infect. 2012; 18:834–840. PMID: 22827611.
Article59. Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008; 105:15064–15069. PMID: 18806221.
Article60. Fung TC, Artis D, Sonnenberg GF. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev. 2014; 260:35–49. PMID: 24942680.
Article61. Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet. 2015; 6:81. PMID: 25852737.
Article62. Schroeder BO, Birchenough GMH, Ståhlman M, et al. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe. 2018; 23:27–40.e7. PMID: 29276171.
Article63. Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014; 111:18321–18326. PMID: 25489084.
Article64. Li S, Konstantinov SR, Smits R, Peppelenbosch MP. Bacterial biofilms in colorectal cancer initiation and progression. Trends Mol Med. 2017; 23:18–30. PMID: 27986421.
Article65. Johnson CH, Dejea CM, Edler D, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015; 21:891–897. PMID: 25959674.
Article66. Drewes JL, Housseau F, Sears CL. Sporadic colorectal cancer: microbial contributors to disease prevention, development and therapy. Br J Cancer. 2016; 115:273–280. PMID: 27380134.
Article67. Mima K, Cao Y, Chan AT, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol. 2016; 7:e200. DOI: 10.1038/ctg.2016.53. PMID: 27811909.
Article68. Fellows R, Denizot J, Stellato C, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun. 2018; 9:105. PMID: 29317660.
Article69. Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014; 40:128–139. PMID: 24412617.
Article70. Hu Y, Le Leu RK, Christophersen CT, et al. Manipulation of the gut microbiota using resistant starch is associated with protection against colitis-associated colorectal cancer in rats. Carcinogenesis. 2016; 37:366–375. PMID: 26905582.
Article71. Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012; 487:104–108. PMID: 22722865.
Article72. Bernstein C, Holubec H, Bhattacharyya AK, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011; 85:863–871. PMID: 21267546.
Article73. Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbialpathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012; 3:448. PMID: 23226130.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gut Microbial Influence and Probiotics on Colorectal Cancer
- The role of microbiome in colorectal carcinogenesis and its clinical potential as a target for cancer treatment
- Crossroad between inflammation and carcinogenesis in colon
- High-fat-diet-modulated Gut Microbiota Promotes Intestinal Carcinogenesis
- Gut Microbiome and Colorectal Cancer