Lineage Differentiation Program of Invariant Natural Killer T Cells
- Affiliations
-
- 1Academy of Immunology and Microbiology, Institute for Basic Science, Pohang 37673, Korea. youjeong77@postech.ac.kr
- 2Division of Integrative Biosciences and Biotechnology, Pohang University of Science and Technology, Pohang 37673, Korea.
- KMID: 2400623
- DOI: http://doi.org/10.4110/in.2017.17.6.365
Abstract
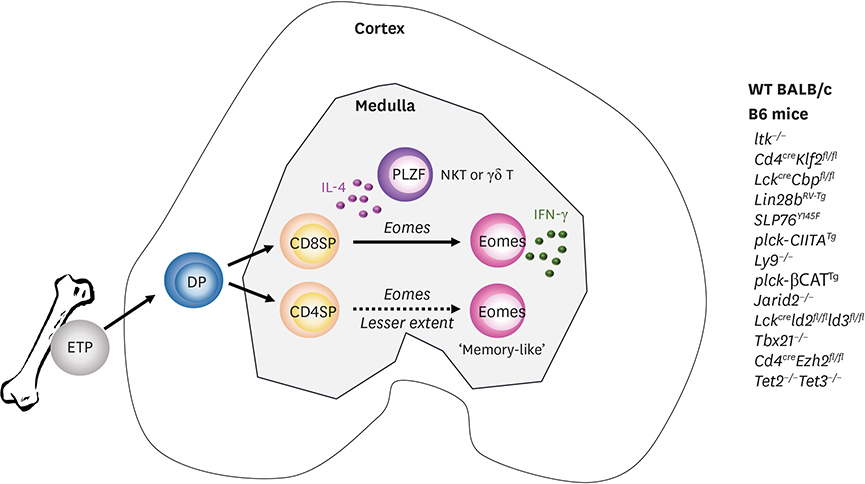
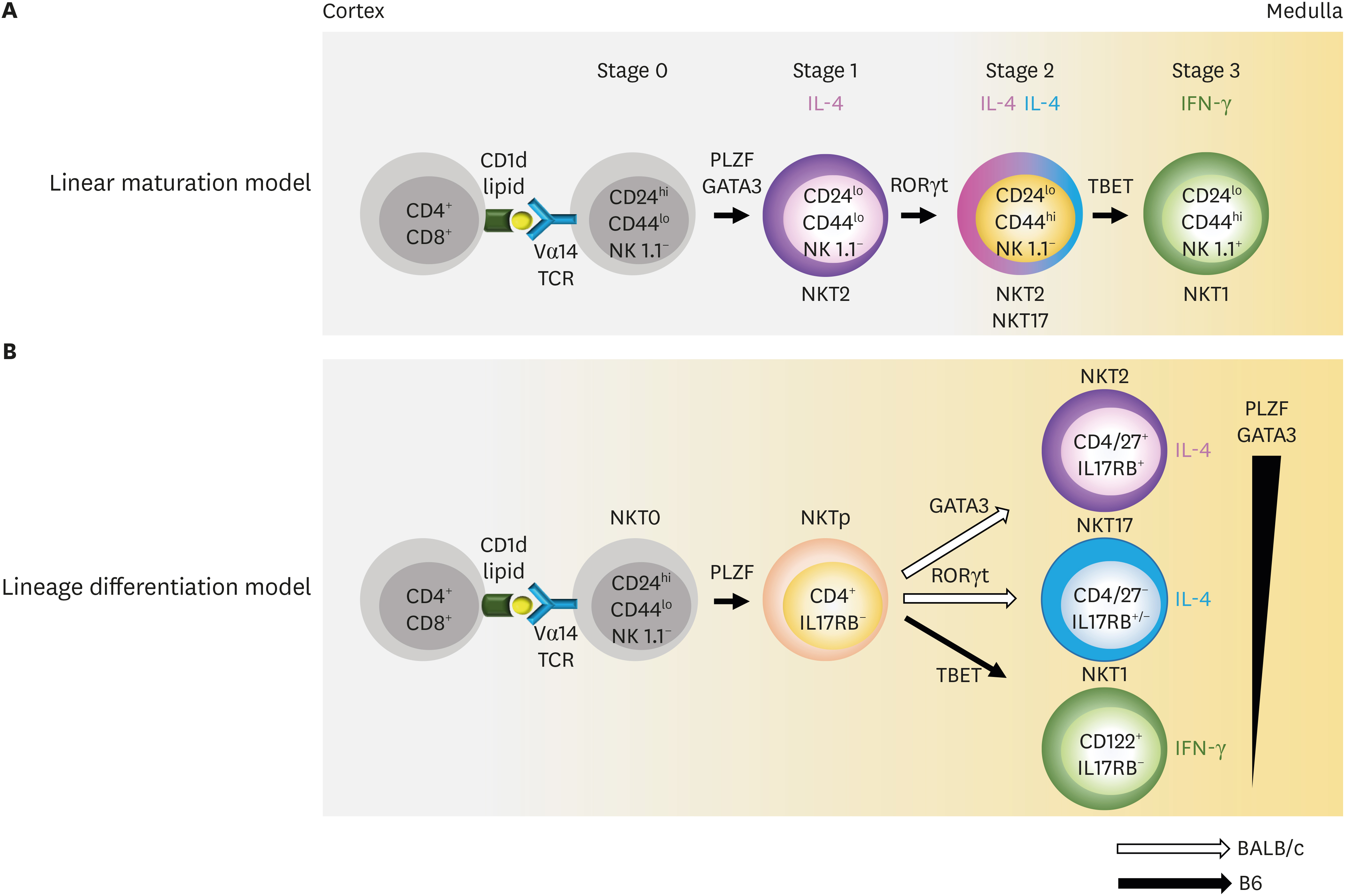
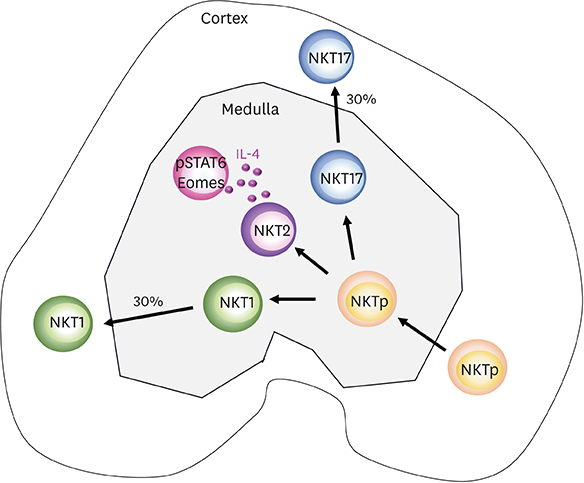
- Invariant natural killer T (iNKT) cells are innate T cells restricted by CD1d molecules. They are positively selected in the thymic cortex and migrate to the medullary area, in which they differentiate into 3 different lineages. Promyelocytic leukemia zinc finger (PLZF) modulates this process, and PLZFhigh, PLZFintermediate, and PLZFlow iNKT cells are designated as NKT2, NKT17, and NKT1 cells, respectively. Analogous to conventional helper CD4 T cells, each subset expresses distinct combinations of transcription factors and produces different cytokines. In lymphoid organs, iNKT subsets have unique localizations, which determine their cytokine responses upon antigenic challenge. The lineage differentiation programs of iNKT cells are differentially regulated in various mice strains in a cell-intrinsic manner, and BALB/c mice contain a high frequency of NKT2 cells. In the thymic medulla, steady state IL-4 from NKT2 cells directly conditions CD8 T cells to become memory-like cells expressing Eomesodermin, which function as premade memory effectors. The genetic signature of iNKT cells is more similar to that of γδ T cells and innate lymphoid cells (ILCs) than of conventional helper T cells, suggesting that ILCs and innate T cells share common developmental programs.
MeSH Terms
Figure
Cited by 6 articles
-
Expansion and Sub-Classification of T Cell-Dependent Antibody Responses to Encompass the Role of Innate-Like T Cells in Antibody Responses
Chanho Park, Tae Jin Kim
Immune Netw. 2018;18(5):. doi: 10.4110/in.2018.18.e34.CD24+ Cell Depletion Permits Effective Enrichment of Thymic iNKT Cells While Preserving Their Subset Composition
Joo-Young Park, Juntae Kwon, Emily Y. Kim, Juliet Fink, Hye Kyung Kim, Jung-Hyun Park
Immune Netw. 2019;19(2):. doi: 10.4110/in.2019.19.e14.Regulation of Allergic Immune Responses by Microbial Metabolites
Hyun Jung Park, Sung Won Lee, Seokmann Hong
Immune Netw. 2018;18(1):e15. doi: 10.4110/in.2018.18.e15.IL-17-Producing Cells in Tumor Immunity: Friends or Foes?
Da-Sol Kuen, Byung-Seok Kim, Yeonseok Chung
Immune Netw. 2020;20(1):e6. doi: 10.4110/in.2020.20.e6.Regulation of Allergic Immune Responses by Microbial Metabolites
Hyun Jung Park, Sung Won Lee, Seokmann Hong
Immune Netw. 2018;18(1):. doi: 10.4110/in.2018.18.e15.IL-17-Producing Cells in Tumor Immunity: Friends or Foes?
Da-Sol Kuen, Byung-Seok Kim, Yeonseok Chung
Immune Netw. 2020;20(1):. doi: 10.4110/in.2020.20.e6.
Reference
-
1. Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. 2015; 16:1114–1123.
Article2. Bedoui S, Gebhardt T, Gasteiger G, Kastenmüller W. Parallels and differences between innate and adaptive lymphocytes. Nat Immunol. 2016; 17:490–494.
Article3. Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005; 22:705–716.
Article4. Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005; 202:239–248.
Article5. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008; 29:391–403.
Article6. Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008; 9:1055–1064.
Article7. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013; 14:1146–1154.
Article8. Lee YJ, Wang H, Starrett GJ, Phuong V, Jameson SC, Hogquist KA. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity. 2015; 43:566–578.
Article9. Lee YJ, Starrett GJ, Lee ST, Yang R, Henzler CM, Jameson SC, Hogquist KA. Lineage-specific effector signatures of invariant NKT cells are shared amongst γδ T, innate lymphoid, and th cells. J Immunol. 2016; 197:1460–1470.
Article10. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007; 25:297–336.
Article11. Sharif S, Arreaza GA, Zucker P, Mi QS, Sondhi J, Naidenko OV, Kronenberg M, Koezuka Y, Delovitch TL, Gombert JM, et al. Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nat Med. 2001; 7:1057–1062.
Article12. Singh AK, Wilson MT, Hong S, Olivares-Villagómez D, Du C, Stanic AK, Joyce S, Sriram S, Koezuka Y, Van Kaer L. Natural killer T cell activation protects mice against experimental autoimmune encephalomyelitis. J Exp Med. 2001; 194:1801–1811.
Article13. Jahng AW, Maricic I, Pedersen B, Burdin N, Naidenko O, Kronenberg M, Koezuka Y, Kumar V. Activation of natural killer T cells potentiates or prevents experimental autoimmune encephalomyelitis. J Exp Med. 2001; 194:1789–1799.
Article14. Nieuwenhuis EE, Matsumoto T, Exley M, Schleipman RA, Glickman J, Bailey DT, Corazza N, Colgan SP, Onderdonk AB, Blumberg RS. CD1d-dependent macrophage-mediated clearance of Pseudomonas aeruginosa from lung. Nat Med. 2002; 8:588–593.
Article15. Morshed SR, Mannoor K, Halder RC, Kawamura H, Bannai M, Sekikawa H, Watanabe H, Abo T. Tissue-specific expansion of NKT and CD5+B cells at the onset of autoimmune disease in (NZBxNZW)F1 mice. Eur J Immunol. 2002; 32:2551–2561.
Article16. Akbari O, Stock P, Meyer E, Kronenberg M, Sidobre S, Nakayama T, Taniguchi M, Grusby MJ, DeKruyff RH, Umetsu DT. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003; 9:582–588.
Article17. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. 2002; 17:629–638.
Article18. Kim HY, Kim HJ, Min HS, Kim S, Park WS, Park SH, Chung DH. NKT cells promote antibody-induced joint inflammation by suppressing transforming growth factor beta1 production. J Exp Med. 2005; 201:41–47.
Article19. Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002; 3:772–779.
Article20. Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006; 25:93–104.
Article21. Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006; 25:79–91.
Article22. Fukuyama T, Kasper LH, Boussouar F, Jeevan T, van Deursen J, Brindle PK. Histone acetyltransferase CBP is vital to demarcate conventional and innate CD8+ T-cell development. Mol Cell Biol. 2009; 29:3894–3904.
Article23. Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007; 7:479–485.
Article24. Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010; 11:709–716.
Article25. Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009; 31:122–130.
Article26. Tsagaratou A, González-Avalos E, Rautio S, Scott-Browne JP, Togher S, Pastor WA, Rothenberg EV, Chavez L, Lähdesmäki H, Rao A. TET proteins regulate the lineage specification and TCR-mediated expansion of iNKT cells. Nat Immunol. 2017; 18:45–53.
Article27. Jameson SC, Lee YJ, Hogquist KA. Innate memory T cells. Adv Immunol. 2015; 126:173–213.
Article28. Ventre E, Brinza L, Schicklin S, Mafille J, Coupet CA, Marçais A, Djebali S, Jubin V, Walzer T, Marvel J. Negative regulation of NKG2D expression by IL-4 in memory CD8 T cells. J Immunol. 2012; 189:3480–3489.
Article29. Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011; 32:50–56.
Article30. Vasanthakumar A, Xu D, Lun AT, Kueh AJ, van Gisbergen KP, Iannarella N, Li X, Yu L, Wang D, Williams BR, et al. A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 2017; 18:619–631.
Article31. Dobenecker MW, Kim JK, Marcello J, Fang TC, Prinjha R, Bosselut R, Tarakhovsky A. Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J Exp Med. 2015; 212:297–306.
Article32. Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010; 207:237–246.
Article33. Min HS, Lee YJ, Jeon YK, Kim EJ, Kang BH, Jung KC, Chang CH, Park SH. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. J Immunol. 2011; 186:5749–5757.
Article34. Lai D, Zhu J, Wang T, Hu-Li J, Terabe M, Berzofsky JA, Clayberger C, Krensky AM. KLF13 sustains thymic memory-like CD8(+) T cells in BALB/c mice by regulating IL-4-generating invariant natural killer T cells. J Exp Med. 2011; 208:1093–1103.
Article35. D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010; 11:240–249.36. Thapa P, Romero Arocha S, Chung JY, Sant'Angelo DB, Shapiro VS. Histone deacetylase 3 is required for iNKT cell development. Sci Rep. 2017; 7:5784.
Article37. Beyaz S, Kim JH, Pinello L, Xifaras ME, Hu Y, Huang J, Kerenyi MA, Das PP, Barnitz RA, Herault A, et al. The histone demethylase UTX regulates the lineage-specific epigenetic program of invariant natural killer T cells. Nat Immunol. 2017; 18:184–195.
Article38. Morrot A, Hafalla JC, Cockburn IA, Carvalho LH, Zavala F. IL-4 receptor expression on CD8+ T cells is required for the development of protective memory responses against liver stages of malaria parasites. J Exp Med. 2005; 202:551–560.
Article39. Carvalho LH, Sano G, Hafalla JC, Morrot A, Curotto de Lafaille MA, Zavala F. IL-4-secreting CD4+ T cells are crucial to the development of CD8+ T-cell responses against malaria liver stages. Nat Med. 2002; 8:166–170.
Article40. Renkema KR, Lee JY, Lee YJ, Hamilton SE, Hogquist KA, Jameson SC. IL-4 sensitivity shapes the peripheral CD8+ T cell pool and response to infection. J Exp Med. 2016; 213:1319–1329.
Article41. Lee A, Park SP, Park CH, Kang BH, Park SH, Ha SJ, Jung KC. IL-4 induced innate CD8+ T cells control persistent viral infection. PLoS Pathog. 2015; 11:e1005193.
Article42. White JT, Cross EW, Kedl RM. Antigen-inexperienced memory CD8(+) T cells: where they come from and why we need them. Nat Rev Immunol. 2017; 17:391–400.
Article43. Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Curr Opin Immunol. 2013; 25:161–167.
Article44. Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012; 10:e1001255.
Article45. Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity. 2005; 23:419–429.
Article46. Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, Ho IC. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006; 177:6650–6659.
Article47. Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004; 20:477–494.
Article48. Clancy-Thompson E, Chen GZ, Tyler PM, Servos MM, Barisa M, Brennan PJ, Ploegh HL, Dougan SK. Monoclonal invariant NKT (iNKT) cell mice reveal a role for both tissue of origin and the TCR in development of iNKT functional subsets. J Immunol. 2017; 199:159–171.
Article49. Thomas SY, Scanlon ST, Griewank KG, Constantinides MG, Savage AK, Barr KA, Meng F, Luster AD, Bendelac A. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. J Exp Med. 2011; 208:1179–1188.
Article50. King IL, Amiel E, Tighe M, Mohrs K, Veerapen N, Besra G, Mohrs M, Leadbetter EA. The mechanism of splenic invariant NKT cell activation dictates localization in vivo. J Immunol. 2013; 191:572–582.
Article51. Gray EE, Friend S, Suzuki K, Phan TG, Cyster JG. Subcapsular sinus macrophage fragmentation and CD169+ bleb acquisition by closely associated IL-17-committed innate-like lymphocytes. PLoS One. 2012; 7:e38258.
Article52. Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB. ImmGen Project Consortium. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013; 14:90–99.
Article53. Mycko MP, Ferrero I, Wilson A, Jiang W, Bianchi T, Trumpp A, MacDonald HR. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J Immunol. 2009; 182:4641–4648.
Article54. Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci USA. 2009; 106:8641–8646.
Article55. Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. 2016; 17:728–739.
Article56. Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M. Immunological Genome Consortium. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015; 16:306–317.
Article57. Stubbington MJ, Mahata B, Svensson V, Deonarine A, Nissen JK, Betz AG, Teichmann SA. An atlas of mouse CD4(+) T cell transcriptomes. Biol Direct. 2015; 10:14.
Article58. Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, Kornfeld H, Xiong N, Cohen NR, Brenner MB, et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol. 2012; 13:511–518.
Article59. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014; 508:397–401.
Article60. Strong BS, Newkold TJ, Lee AE, Turner LE, Alhajjat AM, Heusel JW, Shaaban AF. Extrinsic allospecific signals of hematopoietic origin dictate iNKT cell lineage-fate decisions during development. Sci Rep. 2016; 6:28837.
Article61. Haas JD, Ravens S, Düber S, Sandrock I, Oberdörfer L, Kashani E, Chennupati V, Föhse L, Naumann R, Weiss S, et al. Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity. 2012; 37:48–59.
Article62. Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012; 335:1195–1200.
Article63. Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM, Kimura MY, Sharrow SO, Guinter TI, Feigenbaum L, et al. Let-7 microRNAs target the lineage-specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol. 2015; 16:517–524.
Article64. Chang PP, Barral P, Fitch J, Pratama A, Ma CS, Kallies A, Hogan JJ, Cerundolo V, Tangye SG, Bittman R, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2011; 13:35–43.
Article65. Lynch L, Michelet X, Zhang S, Brennan PJ, Moseman A, Lester C, Besra G, Vomhof-Dekrey EE, Tighe M, Koay HF, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of T(reg) cells and macrophages in adipose tissue. Nat Immunol. 2015; 16:85–95.
Article66. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014; 124:3725–3740.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Intracellular Bacterial Infection and Invariant NKT Cells
- Comparison of Invariant NKT Cells with Conventional T Cells by Using Gene Set Enrichment Analysis (GSEA)
- Natural killer T cell and pathophysiology of asthma
- Increased Th2-like Invariant Natural Killer T cells in Peripheral Blood From Patients With Asthma
- Unusual Suspects in the Development of Obesity-Induced Inflammation and Insulin Resistance: NK cells, iNKT cells, and ILCs