Yonsei Med J.
2014 Mar;55(2):324-330.
Small Double-Stranded RNA Mediates the Anti-Cancer Effects of p21(WAF1/ClP1) Transcriptional Activation in a Human Glioma Cell Line
- Affiliations
-
- 1Lanzhou University, Lanzhou, China.
- 2The First Clinical Medical College of Lanzhou University, Lanzhou, China.
- 3Department of Urology, People's Hospital of Gansu Province, Lanzhou, China. chenyirong1688@163.com
Abstract
- PURPOSE
This study was conducted to investigate the small double-stranded RNA (dsRNA) mediated anti-tumor effects of p21(WAF1/ClP1) (p21) transcriptional activation in vitro in the human glioma SHG-44 cell line.
MATERIALS AND METHODS
Human glioma SHG-44 cells were transfected with dsRNA using LipofectAMINE 2000 transfection reagent. Real-time PCR and Western blot analysis were conducted to detect p21 and survivin mRNA and protein levels, respectively. Cell proliferation was examined by MTT assay. Cell cycle distribution and apoptosis were detected by flow-cytometric analysis.
RESULTS
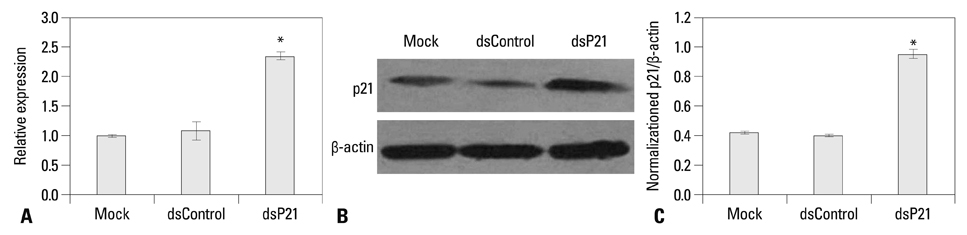
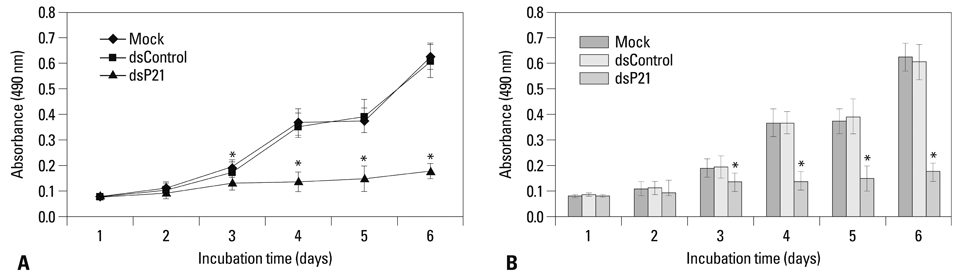
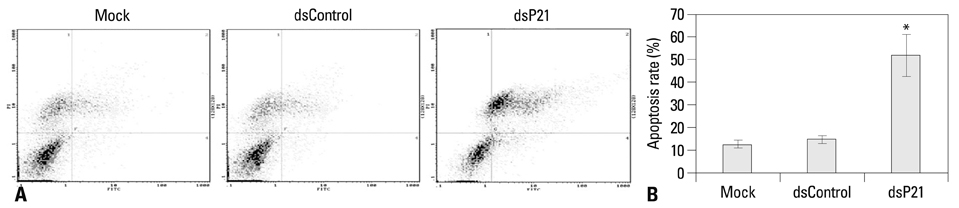
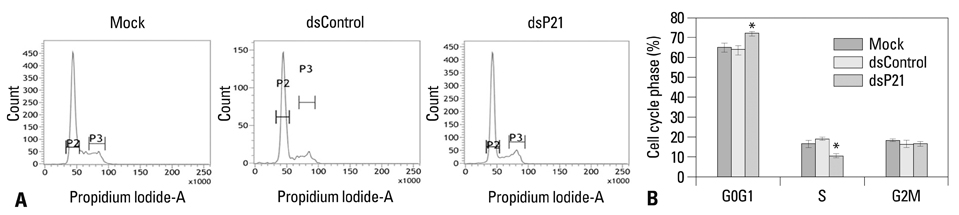
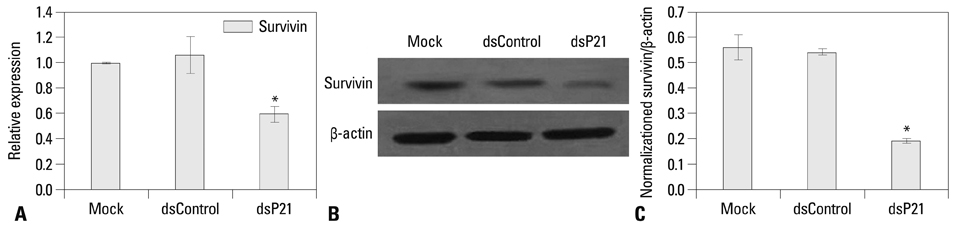
We found that dsRNA targeting p21 promoter (dsP21) significantly induced the expression of p21 at transcription and protein levels, and reduced the expression of survivin. AS well, dsP21 transcription significantly inhibited human glioma SHG-44 cell proliferation. Analysis of cell cycle distribution revealed that dsP21 transfection increased accumulation of cells in the G0/G1 phase and reduced accumulation of cells in the S phase. Further analysis revealed that dsP21 transcription led to an increase in both early and late stages of apoptosis in human glioma SHG-44 cells.
CONCLUSION
In the present study, P21 activation by RNA-induced gene activation (RNAa) induced anti-tumor activity in vitro in a human glioma SHG-44 cell line. The results suggested that RNAa could be used for human glioma treatment by targeted activation of tumor suppressor genes.
Keyword
MeSH Terms
Figure
Reference
-
1. Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004; 64:6892–6899.2. Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006; 103:17337–17342.
Article3. Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, et al. RNAa is conserved in mammalian cells. PLoS One. 2010; 5:e8848.
Article4. Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008; 105:1608–1613.
Article5. Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004; 116:779–793.
Article6. Portnoy V, Huang V, Place RF, Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011; 2:748–760.
Article7. Junxia W, Ping G, Yuan H, Lijun Z, Jihong R, Fang L, et al. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer Sci. 2010; 101:1790–1796.
Article8. Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995; 55:5187–5190.9. Shi YZ, Hui AM, Takayama T, Li X, Cui X, Makuuchi M. Reduced p21(WAF1/CIP1) protein expression is predominantly related to altered p53 in hepatocellular carcinomas. Br J Cancer. 2000; 83:50–55.
Article10. Goto M, Tsukamoto T, Inada K, Mizoshita T, Ogawa T, Terada A, et al. Loss of p21WAF1/CIP1 expression in invasive fronts of oral tongue squamous cell carcinomas is correlated with tumor progression and poor prognosis. Oncol Rep. 2005; 14:837–846.
Article11. Shoji T, Tanaka F, Takata T, Yanagihara K, Otake Y, Hanaoka N, et al. Clinical significance of p21 expression in non-small-cell lung cancer. J Clin Oncol. 2002; 20:3865–3871.
Article12. Wei J, Zhao J, Long M, Han Y, Wang X, Lin F, et al. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer. 2010; 10:632.
Article13. Whitson JM, Noonan EJ, Pookot D, Place RF, Dahiya R. Double stranded-RNA-mediated activation of P21 gene induced apoptosis and cell cycle arrest in renal cell carcinoma. Int J Cancer. 2009; 125:446–452.
Article14. Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol Cancer Ther. 2008; 7:698–703.
Article15. Wu ZM, Dai C, Huang Y, Zheng CF, Dong QZ, Wang G, et al. Anti-cancer effects of p21WAF1/CIP1 transcriptional activation induced by dsRNAs in human hepatocellular carcinoma cell lines. Acta Pharmacol Sin. 2011; 32:939–946.
Article16. Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997; 3:917–921.
Article17. Suzuki A, Shiraki K. Tumor cell "dead or alive": caspase and survivin regulate cell death, cell cycle and cell survival. Histol Histopathol. 2001; 16:583–593.18. Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, et al. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000; 19:1346–1353.
Article19. Zhang Z, Li D, Wu M, Xiang B, Wang L, Zhou M, et al. Promoter hypermethylation-mediated inactivation of LRRC4 in gliomas. BMC Mol Biol. 2008; 9:99.
Article20. Ferrandina G, Stoler A, Fagotti A, Fanfani F, Sacco R, De Pasqua A, et al. p21WAF1/CIP1 protein expression in primary ovarian cancer. Int J Oncol. 2000; 17:1231–1235.
Article21. Cheung TH, Lo KW, Yu MM, Yim SF, Poon CS, Chung TK, et al. Aberrant expression of p21(WAF1/CIP1) and p27(KIP1) in cervical carcinoma. Cancer Lett. 2001; 172:93–98.
Article22. Rau B, Sturm I, Lage H, Berger S, Schneider U, Hauptmann S, et al. Dynamic expression profile of p21WAF1/CIP1 and Ki-67 predicts survival in rectal carcinoma treated with preoperative radiochemotherapy. J Clin Oncol. 2003; 21:3391–3401.
Article23. Sarbia M, Gabbert HE. Modern pathology: prognostic parameters in squamous cell carcinoma of the esophagus. Recent Results Cancer Res. 2000; 155:15–27.
Article24. Xia X, Ma Q, Li X, Ji T, Chen P, Xu H, et al. Cytoplasmic p21 is a potential predictor for cisplatin sensitivity in ovarian cancer. BMC Cancer. 2011; 11:399.
Article25. Hukkelhoven E, Liu Y, Yeh N, Ciznadija D, Blain SW, Koff A. Tyrosine phosphorylation of the p21 cyclin-dependent kinase inhibitor facilitates the development of proneural glioma. J Biol Chem. 2012; 287:38523–38530.
Article26. Besson A, Assoian RK, Roberts JM. Regulation of the cytoskeleton: an oncogenic function for CDK inhibitors. Nat Rev Cancer. 2004; 4:948–955.
Article27. Wang J, Place RF, Huang V, Wang X, Noonan EJ, Magyar CE, et al. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010; 70:10182–10191.
Article28. Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol. 2010; 17:1344–1355.
Article29. Chen R, Wang T, Rao K, Yang J, Zhang S, Wang S, et al. Up-regulation of VEGF by small activator RNA in human corpus cavernosum smooth muscle cells. J Sex Med. 2011; 8:2773–2780.
Article30. Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010; 38:7736–7748.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Effect of Transfer of p21/WAF1 Gene on Kidney and Bladder Cancer Cell Lines
- Expression of Cyclin-dependent Kinase Inhibitor p21(WAF1/CIP1) in Non-small Cell Lung Carcinomas: Relationship with p53 Status and Proliferative Activity
- Chlorpromazine activates p21(Waf1/Cip1) gene transcription via early growth response-1 (Egr-1) in C6 glioma cells
- Differential modulation of zinc-stimulated p21(Cip/WAF1) and cyclin D1 induction by inhibition of PI3 kinase in HT-29 colorectal cancer cells
- Expression of p34(cdc2), p27(Kip1), p21(WAF1/Cip1) and p53 in Human Breast Cancers