Nutr Res Pract.
2015 Aug;9(4):358-363. 10.4162/nrp.2015.9.4.358.
A lifelong exposure to a Western-style diet, but not aging, alters global DNA methylation in mouse colon
- Affiliations
-
- 1Chaum Life Center, CHA University School of Medicine, 442, Dosan-daero, Gangnam-gu, Seoul 135-948, Korea. sang.choi@cha.ac.kr
- 2Friedman School of Nutrition Science and Policy Tufts University, Boston, Massachusetts, USA.
- 3School of Public Health and Health Sciences, University of Massachusetts, Amherst, Massachusetts, USA.
- 4University of Verona School of Medicine, Verona, Italy.
- KMID: 2313851
- DOI: http://doi.org/10.4162/nrp.2015.9.4.358
Abstract
- BACKGROUND/OBJECTIVES
Previous studies have indicated that when compared to young mice, old mice have lower global DNA methylation and higher p16 promoter methylation in colonic mucosa, which is a common finding in colon cancer. It is also known that a Western-style diet (WSD) high in fat and calories, and low in calcium, vitamin D, fiber, methionine and choline (based on the AIN 76A diet) is tumorigenic in colons of mice. Because DNA methylation is modifiable by diet, we investigate whether a WSD disrupts DNA methylation patterns, creating a tumorigenic environment. SUBJECTVIES/METHODS: We investigated the effects of a WSD and aging on global and p16 promoter DNA methylation in the colon. Two month old male C57BL/6 mice were fed either a WSD or a control diet (AIN76A) for 6, 12 or 17 months. Global DNA methylation, p16 promoter methylation and p16 expression were determined by LC/MS, methyl-specific PCR and real time RT-PCR, respectively.
RESULTS
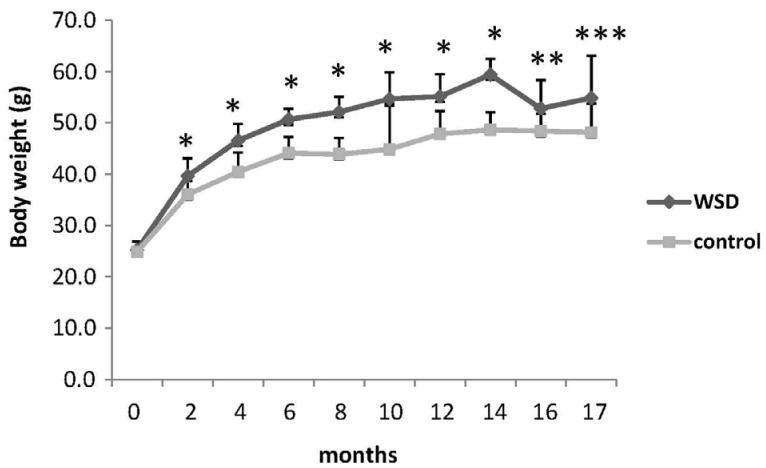
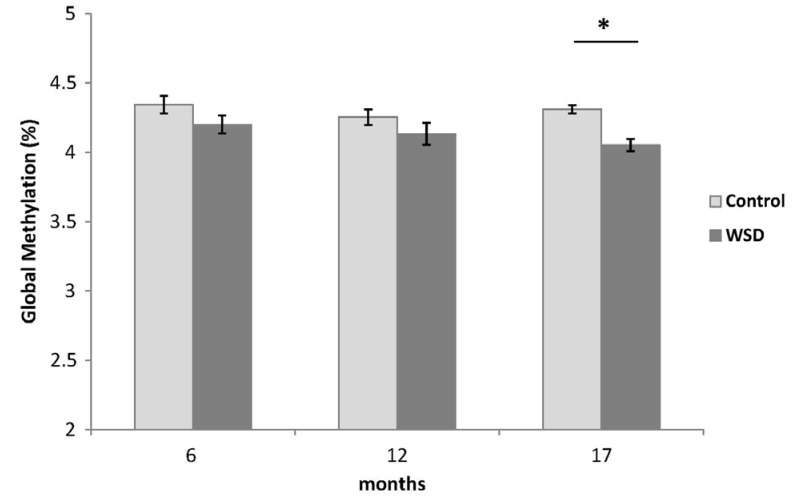
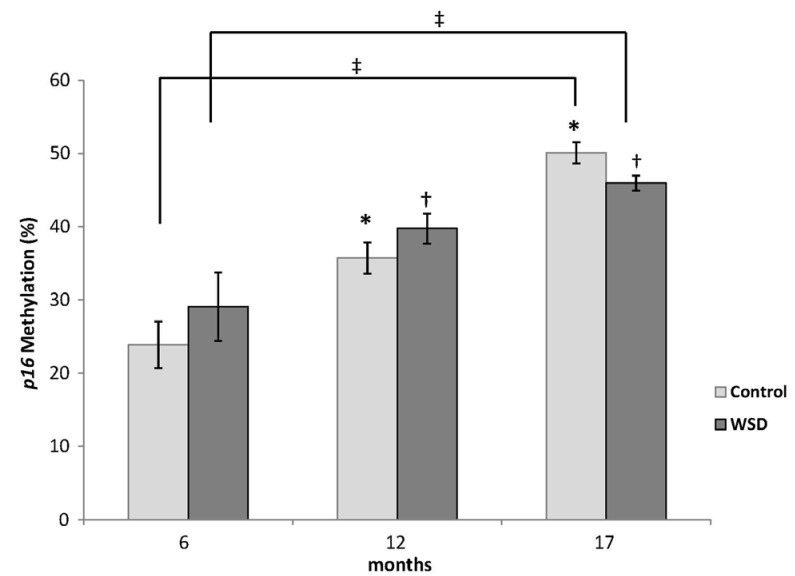
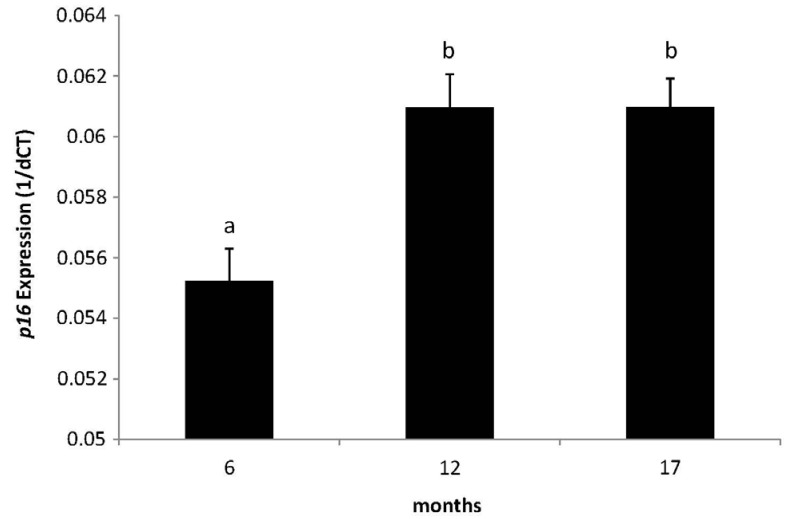
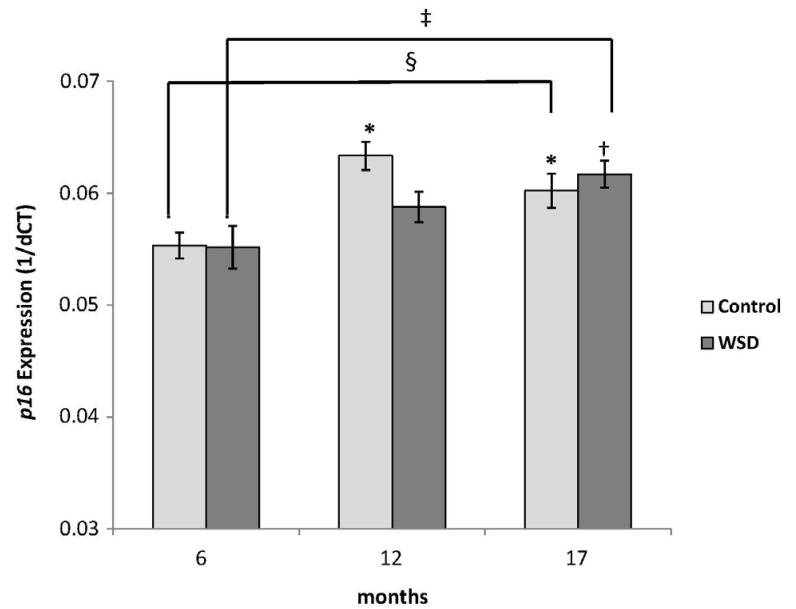
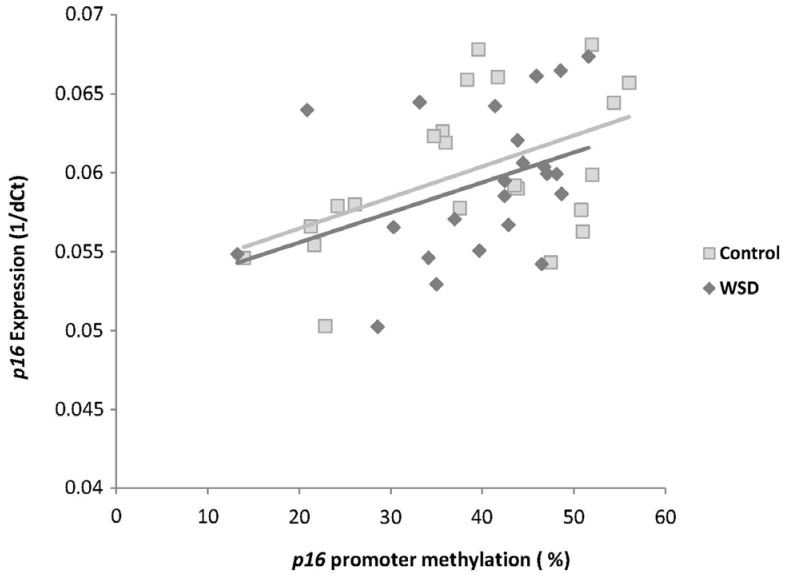
The WSD group demonstrated significantly decreased global DNA methylation compared with the control at 17 months (4.05 vs 4.31%, P = 0.019). While both diets did not change global DNA methylation over time, mice fed the WSD had lower global methylation relative to controls when comparing all animals (4.13 vs 4.30%, P = 0.0005). There was an increase in p16 promoter methylation from 6 to 17 months in both diet groups (P < 0.05) but no differences were observed between diet groups. Expression of p16 increased with age in both control and WSD groups.
CONCLUSIONS
In this model a WSD reduces global DNA methylation, whereas aging itself has no affect. Although the epigenetic effect of aging was not strong enough to alter global DNA methylation, changes in promoter-specific methylation and gene expression occurred with aging regardless of diet, demonstrating the complexity of epigenetic patterns.
Keyword
MeSH Terms
Figure
Reference
-
1. Choi SW, Friso S, Keyes MK, Mason JB. Folate supplementation increases genomic DNA methylation in the liver of elder rats. Br J Nutr. 2005; 93:31–35. PMID: 15705222.
Article2. Keyes MK, Jang H, Mason JB, Liu Z, Crott JW, Smith DE, Friso S, Choi SW. Older age and dietary folate are determinants of genomic and p16-specific DNA methylation in mouse colon. J Nutr. 2007; 137:1713–1717. PMID: 17585020.
Article3. Sauer J, Jang H, Zimmerly EM, Kim KC, Liu Z, Chanson A, Smith DE, Mason JB, Friso S, Choi SW. Ageing, chronic alcohol consumption and folate are determinants of genomic DNA methylation, p16 promoter methylation and the expression of p16 in the mouse colon. Br J Nutr. 2010; 104:24–30. PMID: 20205967.4. Park LK, Friso S, Choi SW. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc. 2012; 71:75–83. PMID: 22051144.
Article5. Newmark HL, Yang K, Lipkin M, Kopelovich L, Liu Y, Fan K, Shinozaki H. A Western-style diet induces benign and malignant neoplasms in the colon of normal C57Bl/6 mice. Carcinogenesis. 2001; 22:1871–1875. PMID: 11698351.
Article6. Yang K, Yang W, Mariadason J, Velcich A, Lipkin M, Augenlicht L. Dietary components modify gene expression: implications for carcinogenesis. J Nutr. 2005; 135:2710–2714. PMID: 16251635.
Article7. Choi SW, Friso S. Epigenetics: a new bridge between nutrition and health. Adv Nutr. 2010; 1:8–16. PMID: 22043447.
Article8. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012; 130:1715–1725. PMID: 22025288.
Article9. Kriegl L, Neumann J, Vieth M, Greten FR, Reu S, Jung A, Kirchner T. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol. 2011; 24:1015–1022. PMID: 21423154.
Article10. Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013; 75:685–705. PMID: 23140366.
Article11. Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013; 34:753–764. PMID: 22906839.
Article12. Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002; 74:4526–4531. PMID: 12236365.
Article13. Frith CH, Highman B, Burger G, Sheldon WD. Spontaneous lesions in virgin and retired breeder BALB/c and C57BL/6 mice. Lab Anim Sci. 1983; 33:273–286. PMID: 6876733.14. James SJ, Muskhelishvili L. Rates of apoptosis and proliferation vary with caloric intake and may influence incidence of spontaneous hepatoma in C57BL/6 x C3H F1 mice. Cancer Res. 1994; 54:5508–5510. PMID: 7923185.15. Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010; 151:4756–4764. PMID: 20685869.
Article16. Widiker S, Karst S, Wagener A, Brockmann GA. High-fat diet leads to a decreased methylation of the Mc4r gene in the obese BFMI and the lean B6 mouse lines. J Appl Genet. 2010; 51:193–197. PMID: 20453306.17. Milagro FI, Campión J, García-Díaz DF, Goyenechea E, Paternain L, Martínez JA. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem. 2009; 65:1–9. PMID: 19588726.
Article18. Zeisel SH. Metabolic crosstalk between choline/1-carbon metabolism and energy homeostasis. Clin Chem Lab Med. 2013; 51:467–475. PMID: 23072856.
Article19. Wu G, Zhang L, Li T, Lopaschuk G, Vance DE, Jacobs RL. Choline deficiency attenuates body weight gain and improves glucose tolerance in ob/ob mice. J Obes. 2012; 2012:319172. PMID: 22778916.
Article20. Rubio-Aliaga I, Roos B, Sailer M, McLoughlin GA, Boekschoten MV, van Erk M, Bachmair EM, van Schothorst EM, Keijer J, Coort SL, Evelo C, Gibney MJ, Daniel H, Muller M, Kleemann R, Brennan L. Alterations in hepatic one-carbon metabolism and related pathways following a high-fat dietary intervention. Physiol Genomics. 2011; 43:408–416. PMID: 21303933.
Article21. Rawson JB, Sun Z, Dicks E, Daftary D, Parfrey PS, Green RC, Gallinger S, McLaughlin JR, Wang PP, Knight JA, Bapat B. Vitamin D intake is negatively associated with promoter methylation of the Wnt antagonist gene DKK1 in a large group of colorectal cancer patients. Nutr Cancer. 2012; 64:919–928. PMID: 22966878.22. Stefanska B, Salamé P, Bednarek A, Fabianowska-Majewska K. Comparative effects of retinoic acid, vitamin D and resveratrol alone and in combination with adenosine analogues on methylation and expression of phosphatase and tensin homologue tumour suppressor gene in breast cancer cells. Br J Nutr. 2012; 107:781–790. PMID: 21801466.
Article23. Stefanska B, Rudnicka K, Bednarek A, Fabianowska-Majewska K. Hypomethylation and induction of retinoic acid receptor beta 2 by concurrent action of adenosine analogues and natural compounds in breast cancer cells. Eur J Pharmacol. 2010; 638:47–53. PMID: 20447390.
Article24. Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, Shi H, Robertson KD, Munn DH, Liu K. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am J Physiol Gastrointest Liver Physiol. 2012; 302:G1405–G1415. PMID: 22517765.
Article25. Astbury SM, Corfe BM. Uptake and metabolism of the short-chain fatty acid butyrate, a critical review of the literature. Curr Drug Metab. 2012; 13:815–821. PMID: 22571479.26. Sarkar S, Abujamra AL, Loew JE, Forman LW, Perrine SP, Faller DV. Histone deacetylase inhibitors reverse CpG methylation by regulating DNMT1 through ERK signaling. Anticancer Res. 2011; 31:2723–2732. PMID: 21868513.27. Spurling CC, Suhl JA, Boucher N, Nelson CE, Rosenberg DW, Giardina C. The short chain fatty acid butyrate induces promoter demethylation and reactivation of RARbeta2 in colon cancer cells. Nutr Cancer. 2008; 60:692–702. PMID: 18791934.28. Goto T, Mizukami H, Shirahata A, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, Hibi K. Aberrant methylation of the p16 gene is frequently detected in advanced colorectal cancer. Anticancer Res. 2009; 29:275–277. PMID: 19331161.29. Zheng S, Pan YX. Histone modifications, not DNA methylation, cause transcriptional repression of p16 (CDKN2A) in the mammary glands of offspring of protein-restricted rats. J Nutr Biochem. 2011; 22:567–573. PMID: 20934317.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- DNA Methylation in the Hypothalamic Feeding Center and Obesity
- DNA Methylation in Brain and Liver Tissues of Mice Infected with Scrapie Agent
- Effects of β-carotene on Expression of Selected MicroRNAs, Histone Acetylation, and DNA Methylation in Colon Cancer Stem Cells
- DNA Methylation in Development
- DNA Methylation-Based Age Estimation in the Forensic Field