J Breast Cancer.
2016 Jun;19(2):133-141. 10.4048/jbc.2016.19.2.133.
Aberrant Promoter Methylation at CpG Cytosines Induce the Upregulation of the E2F5 Gene in Breast Cancer
- Affiliations
-
- 1Cancer Genetics and Epigenetics Lab, Department of Biosciences, COMSATS Institute of Information Technology, Islamabad, Pakistan. muhammad.saeed@comsats.edu.pk
- 2Department of Biotechnology & Genetic Engineering, Kohat University of Science & Technology, Kohat, Pakistan.
- 3Department of Surgical C Unit, Post Graduate Medical Institution, Lady Reading Hospital, Peshawar, Pakistan.
- 4Department of Surgery, Divisional Headquarter Hospital, Kohat, Pakistan.
- KMID: 2308963
- DOI: http://doi.org/10.4048/jbc.2016.19.2.133
Abstract
- PURPOSE
The promoter methylation status of cell cycle regulatory genes plays a crucial role in the regulation of the eukaryotic cell cycle. CpG cytosines are actively subjected to methylation during tumorigenesis, resulting in gain/loss of function. E2F5 gene has growth repressive activities; various studies suggest its involvement in tumorigenesis. This study aims to investigate the epigenetic regulation of E2F5 in breast cancer to better understand tumor biology.
METHODS
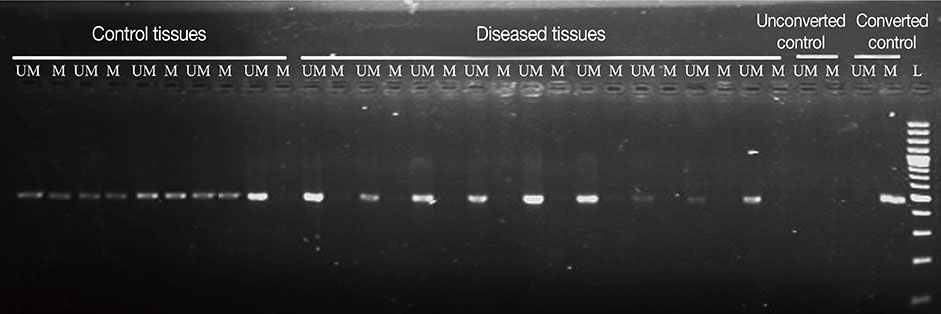
The promoter methylation status of 50 breast tumor tissues and adjacent normal control tissues was analyzed. mRNA expression was determined using SYBR® green quantitative polymerase chain reaction (PCR), and methylation-specific PCR was performed for bisulfite-modified genomic DNA using E2F5-specific primers to assess promoter methylation. Data was statistically analyzed.
RESULTS
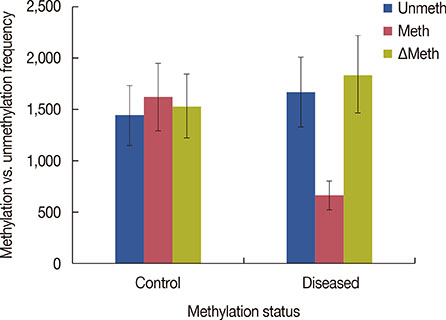
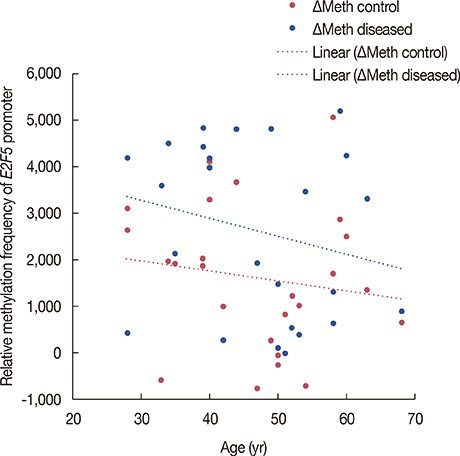
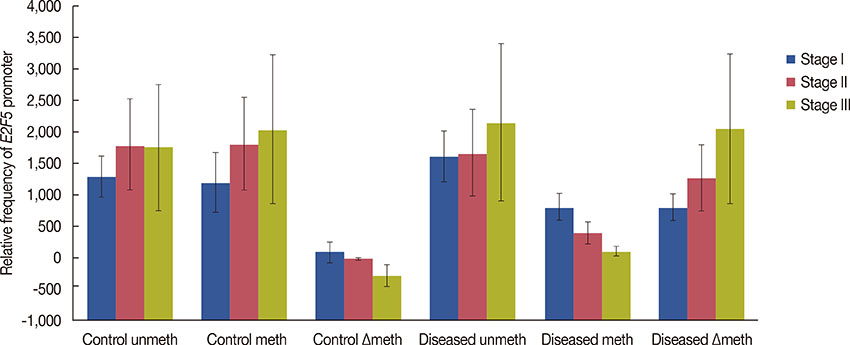
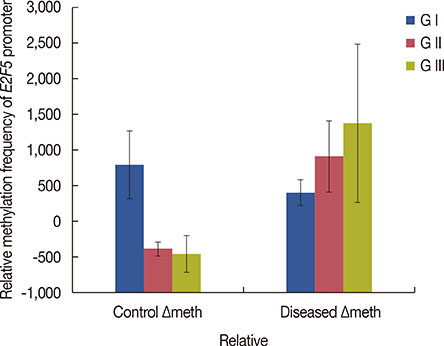
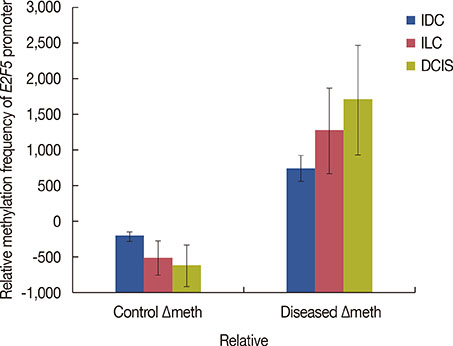
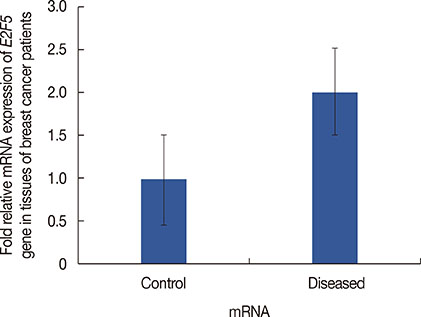
Significant (p<0.001) upregulation was observed in E2F5 expression among tumor tissues, relative to the control group. These samples were hypo-methylated at the E2F5 promoter region in the tumor tissues, compared to the control. Change in the methylation status (Δmeth) was significantly lower (p=0.022) in the tumor samples, indicating possible involvement in tumorigenesis. Patients at the postmenopausal stage showed higher methylation (75%) than those at the premenopausal stage (23.1%). Interestingly, methylation levels gradually increased from the early to the advanced stages of the disease (p<0.001), which suggests a putative role of E2F5 methylation in disease progression that can significantly modulate tumor biology at more advanced stage and at postmenopausal age (Pearson's r=0.99 and 0.86, respectively). Among tissues with different histological status, methylation frequency was higher in invasive lobular carcinoma (80.0%), followed by invasive ductal carcinoma (46.7%) and ductal carcinoma in situ (20.0%).
CONCLUSION
Methylation is an important epigenetic factor that might be involved in the upregulation of E2F5 gene in tumor tissues, which can be used as a prognostic marker for breast cancer.
MeSH Terms
-
Biology
Breast Neoplasms*
Breast*
Carcinogenesis
Carcinoma, Ductal
Carcinoma, Intraductal, Noninfiltrating
Carcinoma, Lobular
Cell Cycle
Disease Progression
DNA
E2F5 Transcription Factor
Epigenomics
Eukaryotic Cells
Genes, Regulator
Humans
Methylation*
Polymerase Chain Reaction
Promoter Regions, Genetic
RNA, Messenger
Up-Regulation*
DNA
E2F5 Transcription Factor
RNA, Messenger
Figure
Reference
-
1. Colozza M, Azambuja E, Cardoso F, Sotiriou C, Larsimont D, Piccart MJ. Proliferative markers as prognostic and predictive tools in early breast cancer: where are we now? Ann Oncol. 2005; 16:1723–1739.
Article2. Eymin B, Gazzeri S. Role of cell cycle regulators in lung carcinogenesis. Cell Adh Migr. 2010; 4:114–123.
Article3. Kothandaraman N, Bajic VB, Brendan PN, Huak CY, Keow PB, Razvi K, et al. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer. 2010; 10:64.
Article4. Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004; 23:4709–4716.
Article5. Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009; 9:785–797.
Article6. Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013; 14:518–528.
Article7. Crijns AP, Fehrmann RS, de Jong S, Gerbens F, Meersma GJ, Klip HG, et al. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med. 2009; 6:e24.
Article8. Fuchs B, Zhang K, Schabel A, Bolander ME, Sarkar G. Identification of twenty-two candidate markers for human osteogenic sarcoma. Gene. 2001; 278:245–252.
Article9. Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl). 2007; 85:293–304.
Article10. Polanowska J, Le Cam L, Orsetti B, Vallés H, Fabbrizio E, Fajas L, et al. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer. 2000; 28:126–130.
Article11. Reimer D, Sadr S, Wiedemair A, Stadlmann S, Concin N, Hofstetter G, et al. Clinical relevance of E2F family members in ovarian cancer: an evaluation in a training set of 77 patients. Clin Cancer Res. 2007; 13:144–151.
Article12. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010; 31:27–36.
Article13. Baxter E, Windloch K, Gannon F, Lee JS. Epigenetic regulation in cancer progression. Cell Biosci. 2014; 4:45.
Article14. Ng JM, Yu J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int J Mol Sci. 2015; 16:2472–2496.
Article15. Ozdemir F, Altinisik J, Karateke A, Coksuer H, Buyru N. Methylation of tumor suppressor genes in ovarian cancer. Exp Ther Med. 2012; 4:1092–1096.
Article16. Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011; 6:479–507.
Article17. Strauss WM. Preparation of genomic DNA from mammalian tissue. Curr Protoc Mol Biol. 2001; Chapter 2:Unit2.2.
Article18. Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002; 18:1427–1431.
Article19. Lee JT, Tsang WH, Chow KL. Simple modifications to standard TRIzol® protocol allow high-yield RNA extraction from cells on resorbable materials. J Biomater Nanobiotechnol. 2011; 2:41–48.20. Kim TM, Yim SH, Shin SH, Xu HD, Jung YC, Park CK, et al. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer. 2008; 123:2808–2815.
Article21. Hijmans EM, Voorhoeve PM, Beijersbergen RL, van't Veer LJ, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995; 15:3082–3089.
Article22. Le Cam L, Polanowska J, Fabbrizio E, Olivier M, Philips A, Ng Eaton E, et al. Timing of cyclin E gene expression depends on the regulated association of a bipartite repressor element with a novel E2F complex. EMBO J. 1999; 18:1878–1890.
Article23. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015; 163:506–519.24. Aziz Z, Sana S, Akram M, Saeed A. Socioeconomic status and breast cancer survival in Pakistani women. J Pak Med Assoc. 2004; 54:448–453.25. Ullah F, Khan T, Ali N, Malik FA, Kayani MA, Shah ST, et al. Promoter methylation status modulate the expression of tumor suppressor (RbL2/p130) gene in breast cancer. PLoS One. 2015; 10:e0134687.
Article26. Almén MS, Nilsson EK, Jacobsson JA, Kalnina I, Klovins J, Fredriksson R, et al. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene. 2014; 548:61–67.
Article27. Weng NP, Levine BL, June CH, Hodes RJ. Regulated expression of telomerase activity in human T lymphocyte development and activation. J Exp Med. 1996; 183:2471–2479.
Article28. Cui H, Kong Y, Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. J Signal Transduct. 2012; 2012:646354.
Article29. Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013; 754:3–29.
Article30. Yu H, Huang YJ, Liu Z, Wang LE, Li G, Sturgis EM, et al. Effects of MDM2 promoter polymorphisms and p53 codon 72 polymorphism on risk and age at onset of squamous cell carcinoma of the head and neck. Mol Carcinog. 2011; 50:697–706.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- CpG Island Hypermethylation in Gastric Carcinoma and Its Premalignant Lesions
- DNA Methylation Changes in Human Cancers
- AKAP12alpha is Associated with Promoter Methylation in Lung Cancer
- Understanding of epigenetics and DNA methylation
- The Characterization of CpG Methylation of ERalpha and ERbeta Gene in the Breast Cancer