Yonsei Med J.
2019 Sep;60(9):842-853. 10.3349/ymj.2019.60.9.842.
Long Intergenic Non-Protein Coding RNA 665 Regulates Viability, Apoptosis, and Autophagy via the MiR-186-5p/MAP4K3 Axis in Hepatocellular Carcinoma
- Affiliations
-
- 1Department of General Surgery, Jinchang Central Hospital, Jinchang, Gansu, China. intersection88@126.com
- 2Department of Ultrasound, Shenzhen Hospital of Southern Medical University, Shenzhen, China.
- KMID: 2457480
- DOI: http://doi.org/10.3349/ymj.2019.60.9.842
Abstract
- PURPOSE
Long intergenic non-protein coding RNA 665 (LINC00665) plays a vital role in the development of cancer. Its function in hepatocellular carcinoma (HCC), however, remains largely unknown.
MATERIALS AND METHODS
The expressions of LINC00665, miR-186-5p, and MAP4K3 were determined by qRT-PCR. Cell viability and apoptosis were evaluated by MTT and flow cytometry, respectively. Autophagic puncta formation was observed by fluorescence microscopy. Bioinformatics analysis, luciferase reporter assay, RNA immunoprecipitation, and RNA pulldown were performed to identify associations among LINC00665, miR-186-5p, and MAP4K3. Western blot was utilized to examine the expressions of MAP4K3, Beclin-1, and LC3. Tumor growth was evaluated in a xenograft model.
RESULTS
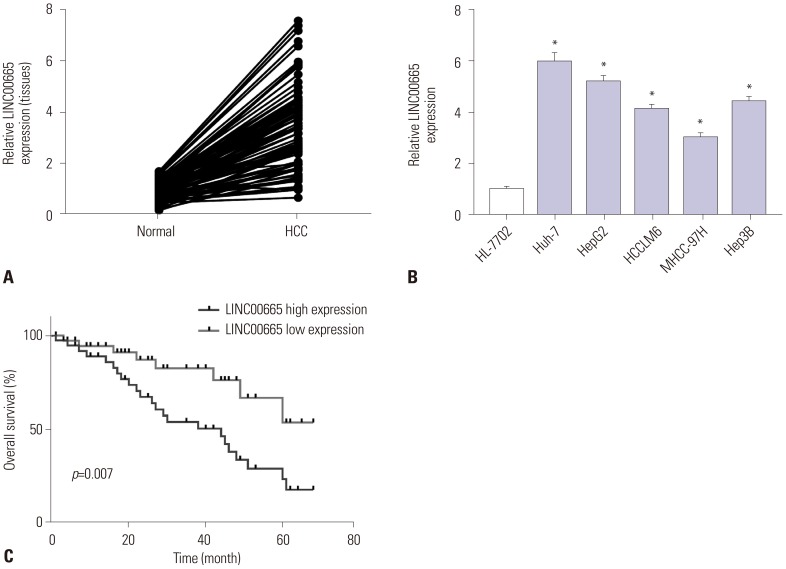
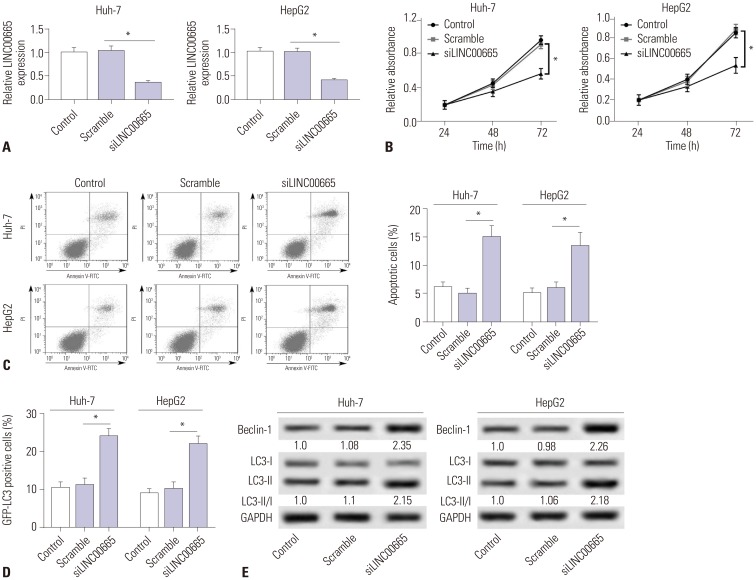
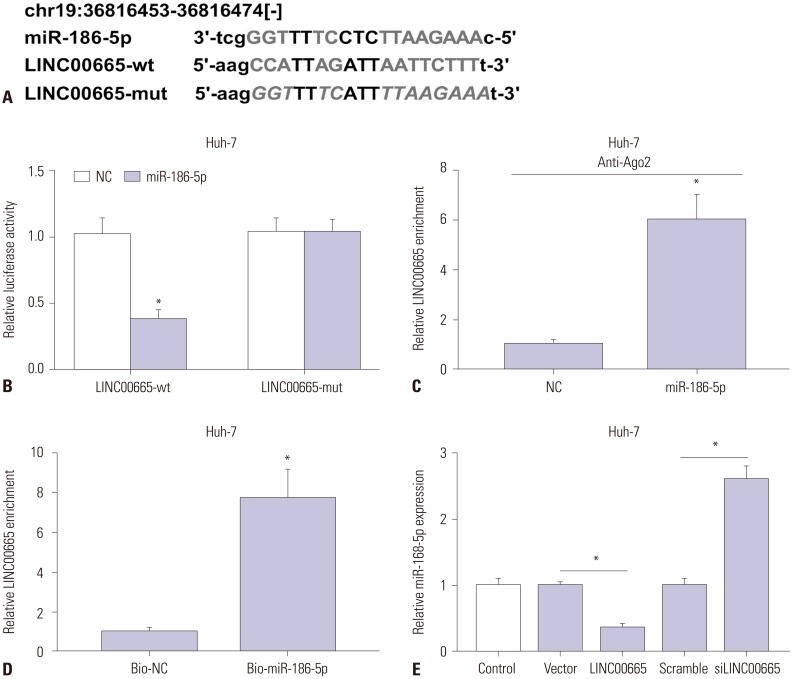
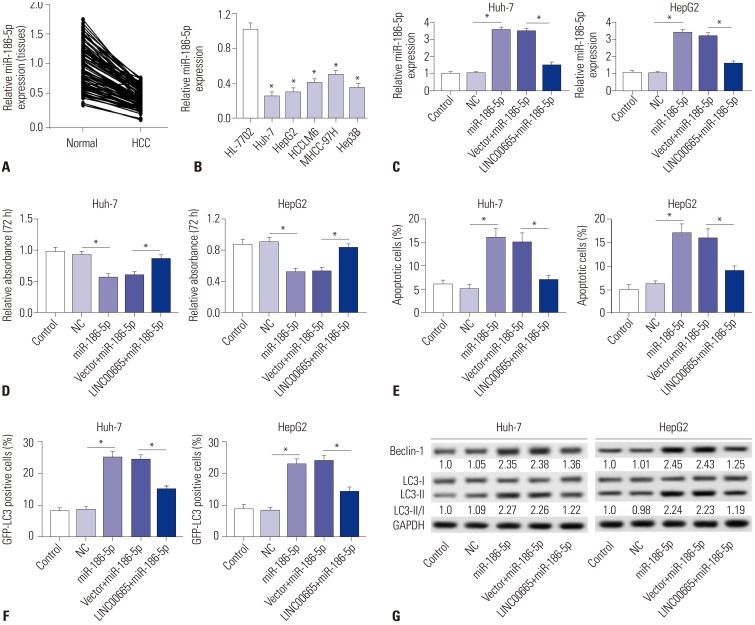
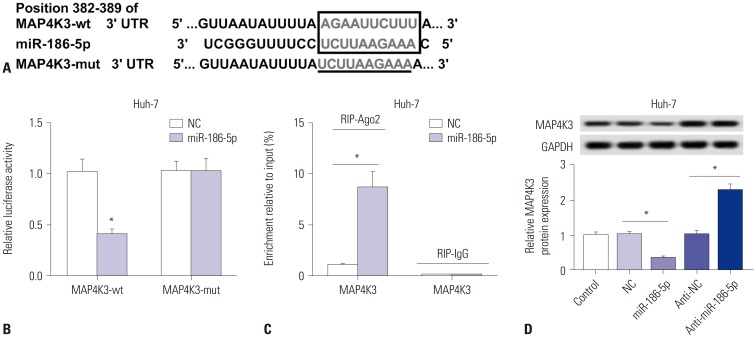
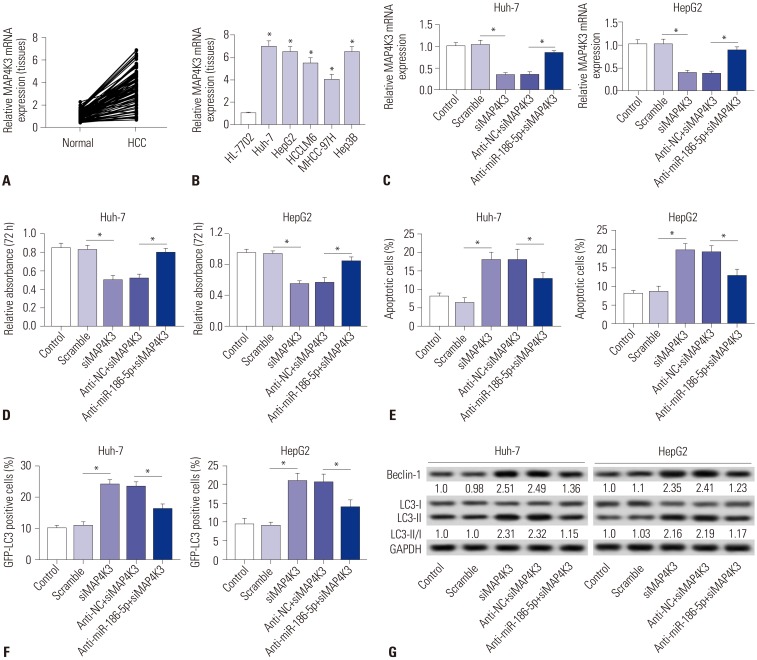
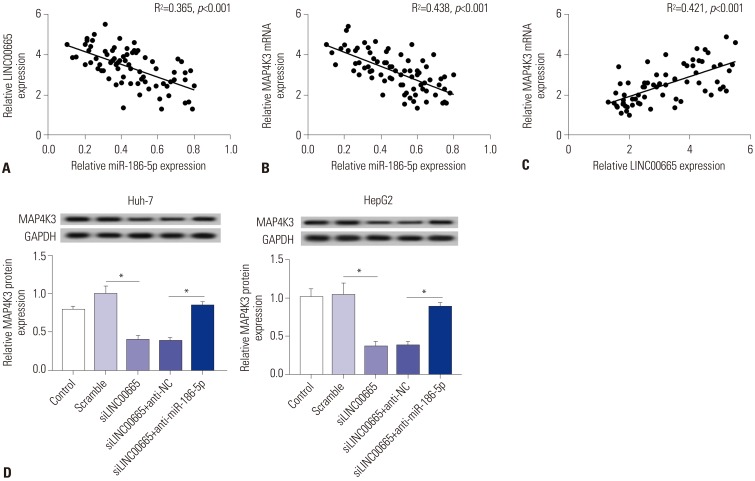
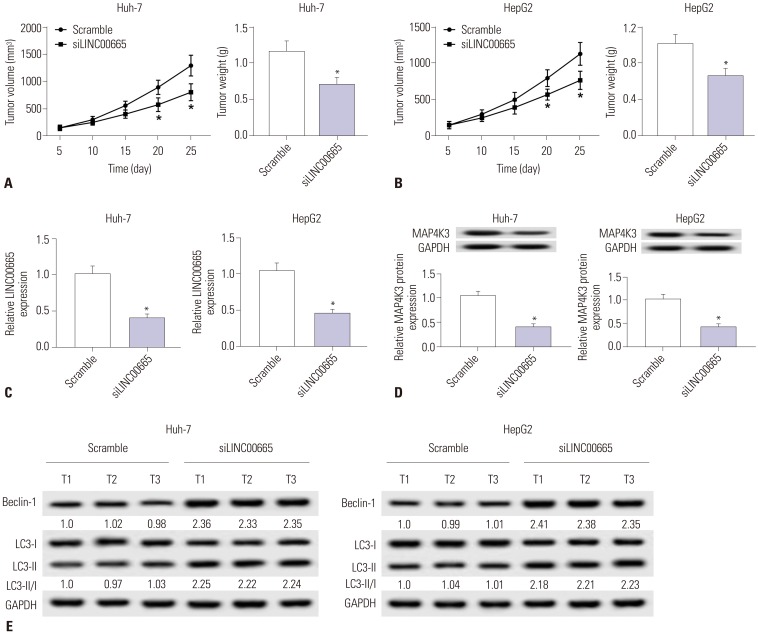
Elevations in LINC00665 were observed in HCC tissues and cells. The overall survival of HCC patients with high levels of LINC00665 was shorter than those with low levels. In vitro, LINC00665 depletion inhibited viability and induced apoptosis and autophagy. miR-186-5p interacted with LINC00665 and was downregulated in HCC tissues and cells. Upregulation of miR-186-5p inhibited viability and induced apoptosis and autophagy, which were attenuated by upregulation of LINC00665. MAP4K3 was found to possess binding sites with miR-186-5p and was upregulated in HCC tissues and cells. MAP4K3 depletion inhibited viability and induced apoptosis and autophagy, which were attenuated by miR-186-5p inhibitor. In vivo, miR-186-5p expression was negatively correlated with LINC00665 or MAP4K3 in HCC tissues, while LINC00665 was positively correlated with MAP4K3. LINC00665 knockdown suppressed tumor growth.
CONCLUSION
LINC00665 was involved in cell viability, apoptosis, and autophagy in HCC via miR-186-5p/MAP4K3 axis, which may provide a new approach for HCC treatment.
Keyword
MeSH Terms
Figure
Reference
-
1. Mak LY, Cruz-Ramón V, Chinchilla-López P, Torres HA, LoConte NK, Rice JP, et al. Global epidemiology, prevention, and management of hepatocellular carcinoma. Am Soc Clin Oncol Educ Book. 2018; 38:262–279. PMID: 30231359.
Article2. Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016; 36:317–324. PMID: 26601627.
Article3. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010; 7:448–458. PMID: 20628345.
Article4. Rastogi A. Changing role of histopathology in the diagnosis and management of hepatocellular carcinoma. World J Gastroenterol. 2018; 24:4000–4013. PMID: 30254404.
Article5. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018; 15:599–616. PMID: 30061739.
Article6. Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol. 2013; 10:1771–1777. PMID: 24384562.
Article7. He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014; 344:20–27. PMID: 24183851.
Article8. Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long noncoding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014; 35:507–514. PMID: 24296588.
Article9. Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014; 2014:780521. PMID: 24757675.
Article10. Sun J, Bie B, Zhang S, Yang J, Li Z. Long non-coding RNAs: critical players in hepatocellular carcinoma. Int J Mol Sci. 2014; 15:20434–20448. PMID: 25387074.11. Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015; 6:7899–7917. PMID: 25760077.
Article12. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015; 75:3181–3191. PMID: 26069248.
Article13. Wen DY, Lin P, Pang YY, Chen G, He Y, Dang YW, et al. Expression of the long intergenic non-protein coding RNA 665 (LINC00665) gene and the cell cycle in hepatocellular carcinoma using The Cancer Genome Atlas, the Gene Expression Omnibus, and Quantitative Real-Time Polymerase Chain Reaction. Med Sci Monit. 2018; 24:2786–2808. PMID: 29728556.
Article14. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014; 9:287–314. PMID: 24079833.
Article15. Song Y, Wang F, Huang Q, Cao Y, Zhao Y, Yang C. MicroRNAs contribute to hepatocellular carcinoma. Mini Rev Med Chem. 2015; 15:459–466. PMID: 25807945.
Article16. Guo W, Tan W, Liu S, Huang X, Lin J, Liang R, et al. MiR-570 inhibited the cell proliferation and invasion through directly targeting B7-H1 in hepatocellular carcinoma. Tumour Biol. 2015; 36:9049–9057. PMID: 26084609.
Article17. Oksuz Z, Serin MS, Kaplan E, Dogen A, Tezcan S, Aslan G, et al. Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p could be used as novel non-invasive biomarkers for HCV-positive cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 2015; 42:713–720. PMID: 25391771.
Article18. Arbogast F, Gros F. Lymphocyte autophagy in homeostasis, activation, and inflammatory diseases. Front Immunol. 2018; 9:1801. PMID: 30127786.
Article19. Wang Z, Choi ME. Autophagy in kidney health and disease. Antioxid Redox Signal. 2014; 20:519–537. PMID: 23642034.
Article20. Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, et al. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018; 37:1142–1158. PMID: 29255248.
Article21. Folkerts H, Hilgendorf S, Vellenga E, Bremer E, Wiersma VR. The multifaceted role of autophagy in cancer and the microenvironment. Med Res Rev. 2019; 39:517–560. PMID: 30302772.
Article22. Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015; 6:8474–8490. PMID: 25893379.
Article23. Zhai H, Fesler A, Ju J. MicroRNA: a third dimension in autophagy. Cell Cycle. 2013; 12:246–250. PMID: 23255136.24. Du F, Feng Y, Fang J, Yang M. MicroRNA-143 enhances chemosensitivity of Quercetin through autophagy inhibition via target GABARAPL1 in gastric cancer cells. Biomed Pharmacother. 2015; 74:169–177. PMID: 26349981.
Article25. Lan T, Yan X, Li Z, Xu X, Mao Q, Ma W, et al. Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186-5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. Tumour Biol. 2017; 39:1010428317705338. PMID: 28656879.
Article26. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015; 36:423–434. PMID: 25968300.
Article27. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014; 25:666–681. PMID: 24768205.
Article28. Huang YH, Al-Aidaroos AQ, Yuen HF, Zhang SD, Shen HM, Rozycka E, et al. A role of autophagy in PTP4A3-driven cancer progression. Autophagy. 2014; 10:1787–1800. PMID: 25136802.
Article29. Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011; 18:571–580. PMID: 21311563.
Article30. Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao XL, et al. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer. 2014; 14:327. PMID: 24885292.
Article31. Hu D, Wu J, Xu L, Zhang R, Chen L. A method for the establishment of a cell line with stable expression of the GFP-LC3 reporter protein. Mol Med Rep. 2012; 6:783–786. PMID: 22825056.
Article32. Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018; 640:53–60. PMID: 29325758.
Article33. Islam F, Gopalan V, Vider J, Wahab R, Ebrahimi F, Lu CT, et al. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017; 357:260–270. PMID: 28549913.
Article34. Wang H, Shen Q, Zhang X, Yang C, Cui S, Sun Y, et al. The long noncoding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017; 41:2221–2229. PMID: 28448993.
Article35. Jones DZ, Schmidt ML, Suman S, Hobbing KR, Barve SS, Gobejishvili L, et al. Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells. BMC Cancer. 2018; 18:421. PMID: 29653561.
Article36. Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011; 75:50–83. PMID: 21372320.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- LncRNA STARD7-AS1 suppresses cervical cancer cell proliferation while promoting autophagy by regulating miR-31-5p/TXNIP axis to inactivate the mTOR signaling
- Long Noncoding RNA Cytoskeleton Regulator RNA Suppresses Apoptosis in Hepatoma Cells by Modulating the miR-125a-5p/ HS1-Associated Protein X-1 Axis to Induce Caspase-9 Inactivation
- Long Non-Coding RNA NORAD Inhibits Breast Cancer Cell Proliferation and Metastasis by Regulating miR-155-5p/ SOCS1 Axis
- Electroacupuncture-Modulated MiR-106b-5p Expression Enhances Autophagy by Targeting Beclin-1 to Promote Motor Function Recovery After Spinal Cord Injury in Rats
- Long Noncoding-RNA Component of Mitochondrial RNA Processing Endoribonuclease Promotes Carcinogenesis in Triple-Negative Breast Cancer Cells via the Competing Endogenous RNA Mechanism