Cancer Res Treat.
2016 Apr;48(2):676-686. 10.4143/crt.2015.153.
Aberrant Epigenetic Modifications of LPHN2 Function as a Potential Cisplatin-Specific Biomarker for Human Gastrointestinal Cancer
- Affiliations
-
- 1Cancer Research Institute, Seoul National University, Seoul, Korea. kimty@snu.ac.kr
- 2Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea.
- 3Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea.
- KMID: 2454346
- DOI: http://doi.org/10.4143/crt.2015.153
Abstract
- PURPOSE
Epigenetic alterations of specific genes have recently been identified as diagnostic biomarkers for human cancers. However, there are currently no standardized epigenetic biomarkers for drug sensitivity in human gastrointestinal cancer. Therefore, the aim of this study is to identify a novel epigenetic biomarker in gastrointestinal cancer.
MATERIALS AND METHODS
Using bisulfite sequencing and pyrosequencing analysis, DNA methylation patterns of gastric, colon primary tissues and their cancer cells were analyzed, and histone modifications were analyzed using chromatin immunoprecipitation assay. In addition, cancer cells were exposed to cisplatin and treated with a DNA methyltransferase inhibitor.
RESULTS
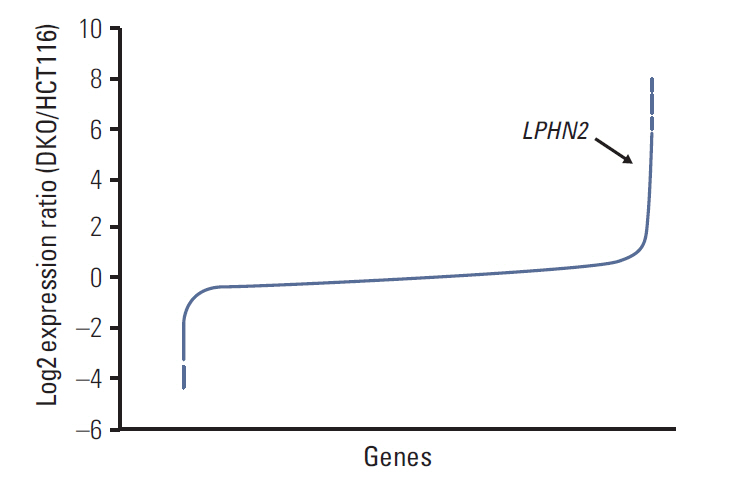
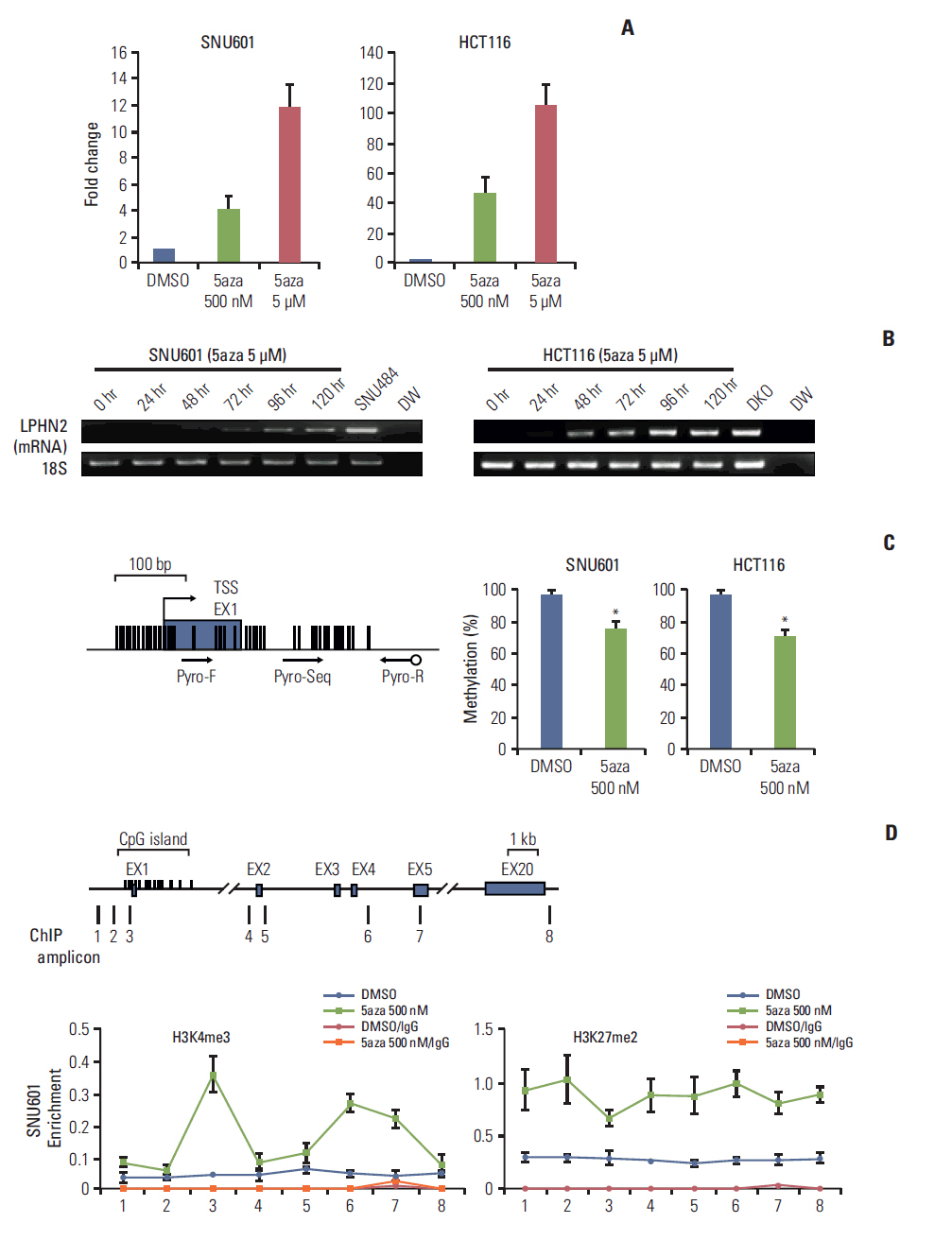
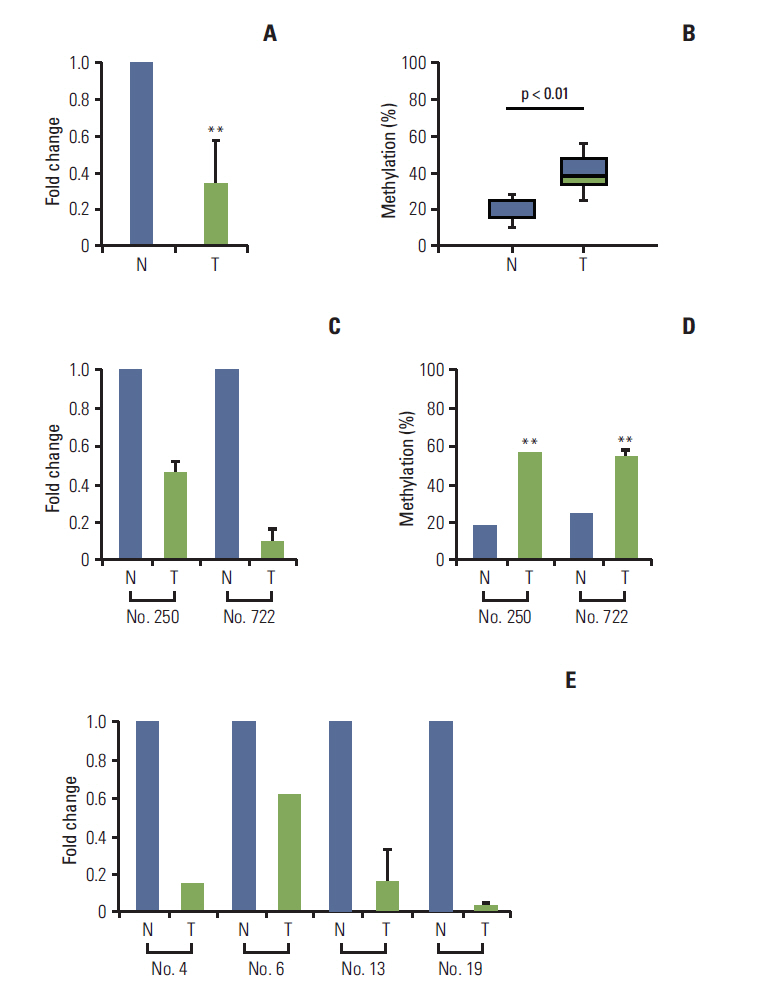
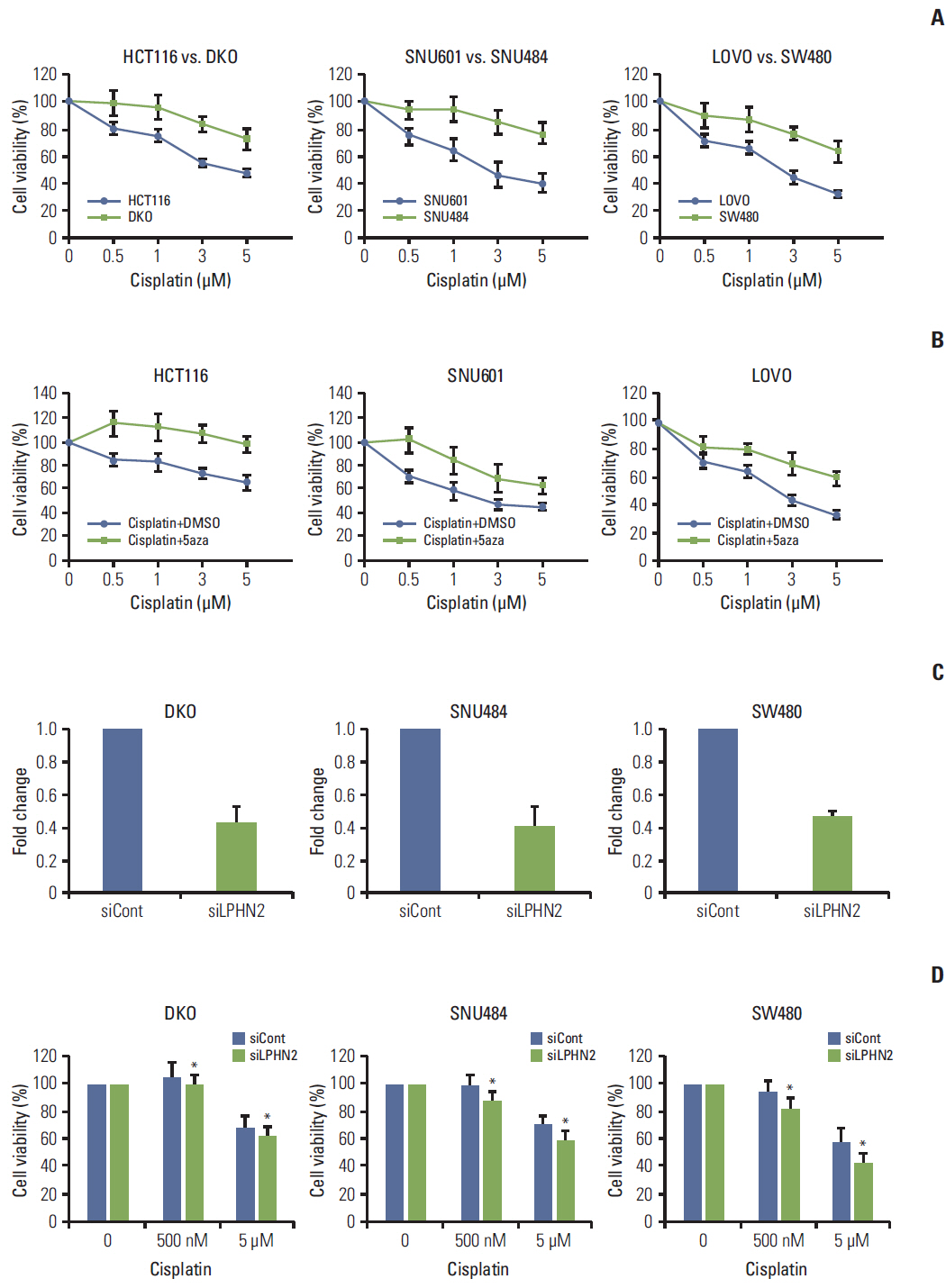
We report that in human gastric and colon cancers, latrophilin 2 (LPHN2) is silenced by epigenetic modifications, including CpG island methylation and aberrant histone modifications. We also confirmed that LPHN2 was silenced by DNA hypermethylation in primary gastric and colon tumor tissues compared to their normal counterparts. Interestingly, we found that cancer cells with methylated LPHN2 showed higher sensitivity to cisplatin. Also, 5-aza- 2"²-deoxycytidine combined with cisplatin decreased the cytotoxicity of cisplatin in cancer cells with methylated LPHN2. In addition, LPHN2 knockdown in cancer cells with high LPHN2 expression sensitized these cells to the anti-proliferative effects of cisplatin.
CONCLUSION
In human gastrointestinal cancer, we found that LPHN2 is regulated by epigenetic modifications, and that cancer cells with lower LPHN2 expression show higher sensitivity to cisplatin. Therefore, the methylation status of LPHN2 is a potential novel epigenetic biomarker for cisplatin treatment in human gastric and colon cancers.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007; 128:683–92.
Article2. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012; 150:12–27.
Article3. Hatziapostolou M, Iliopoulos D. Epigenetic aberrations during oncogenesis. Cell Mol Life Sci. 2011; 68:1681–702.
Article4. Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004; 22:4632–42.
Article5. Jung KW, Won YJ, Oh CM, Kong HJ, Cho H, Lee DH, et al. Prediction of cancer incidence and mortality in Korea, 2015. Cancer Res Treat. 2015; 47:142–8.
Article6. Hamilton JP, Meltzer SJ. A review of the genomics of gastric cancer. Clin Gastroenterol Hepatol. 2006; 4:416–25.
Article7. Nakajima T, Enomoto S, Ushijima T. DNA methylation: a marker for carcinogen exposure and cancer risk. Environ Health Prev Med. 2008; 13:8–15.
Article8. Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, Chernyshov SV, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium Human-Methylation450 BeadChips. Epigenetics. 2013; 8:921–34.
Article9. Fang JY, Xiao SD. Alteration of DNA methylation in gastrointestinal carcinogenesis. J Gastroenterol Hepatol. 2001; 16:960–8.
Article10. Takeshima H, Wakabayashi M, Hattori N, Yamashita S, Ushijima T. Identification of coexistence of DNA methylation and H3K27me3 specifically in cancer cells as a promising target for epigenetic therapy. Carcinogenesis. 2015; 36:192–201.
Article11. Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF, et al. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem. 1997; 272:21504–8.12. Ku JL, Park JG. Biology of SNU cell lines. Cancer Res Treat. 2005; 37:1–19.
Article13. Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002; 416:552–6.
Article14. Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood. 2010; 116:2356–64.15. Yun J, Song SH, Park J, Kim HP, Yoon YK, Lee KH. Gene silencing of EREG mediated by DNA methylation and histone modification in human gastric cancers. Lab Invest. 2012; 92:1033–44.16. Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007; 28:810–22.17. Silva JP, Ushkaryov YA. The latrophilins, "split-personality" receptors. Adv Exp Med Biol. 2010; 706:59–75.
Article18. Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011; 17:330–9.
Article19. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007; 8:286–98.
Article20. Rodriguez J, Munoz M, Vives L, Frangou CG, Groudine M, Peinado MA. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci USA. 2008; 105:19809–14.
Article21. Garritano S, Inga A, Gemignani F, Landi S. More targets, more pathways and more clues for mutant p53. Oncogenesis. 2013; 2:e54.
Article22. Zhang S, Liu Y, Liu Z, Zhang C, Cao H, Ye Y, et al. Transcriptome profiling of a multiple recurrent muscle-invasive urothelial carcinoma of the bladder by deep sequencing. PLoS One. 2014; 9:e91466.
Article23. Zheng CX, Gu ZH, Han B, Zhang RX, Pan CM, Xiang Y, et al. Whole-exome sequencing to identify novel somatic mutations in squamous cell lung cancers. Int J Oncol. 2013; 43:755–64.
Article24. Park KS, Kim HK, Lee JH, Choi YB, Park SY, Yang SH, et al. Transglutaminase 2 as a cisplatin resistance marker in non-small cell lung cancer. J Cancer Res Clin Oncol. 2010; 136:493–502.
Article25. Ivanova T, Zouridis H, Wu Y, Cheng LL, Tan IB, Gopalakrishnan V, et al. Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer. Gut. 2013; 62:22–33.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Epigenetic Cross-Talk between DNA Methylation and Histone Modifications in Human Cancers
- Genetic and epigenetic alterations of colorectal cancer
- Epigenetic Modifications: Novel Therapeutic Approach for Thyroid Cancer
- Epigenetic Alterations and Loss of Imprinting in Colorectal Cancer
- Annexin A5 as a New Potential Biomarker for Cisplatin-Induced Toxicity in Human Kidney Epithelial Cells