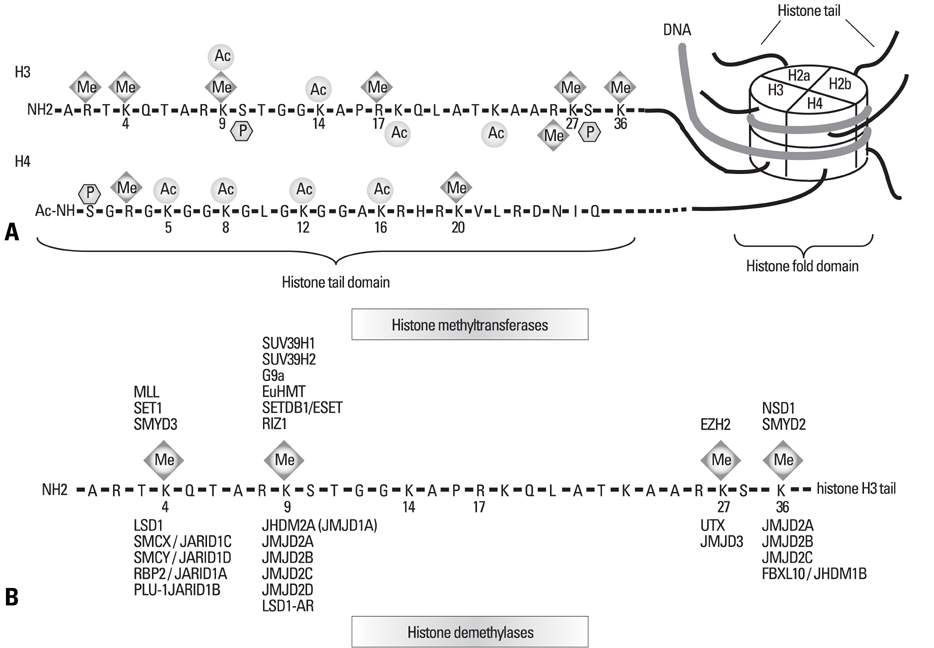
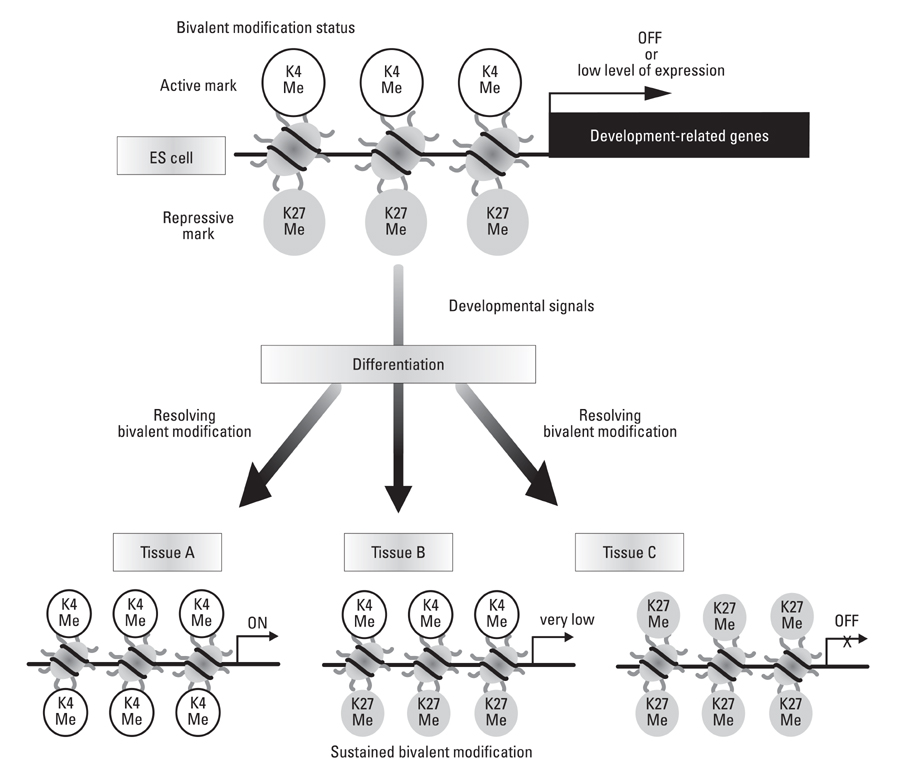
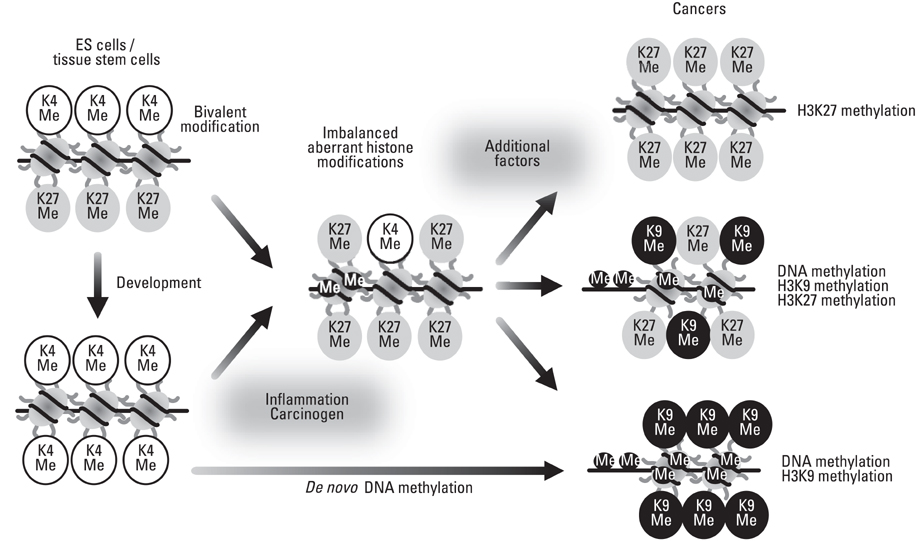
Yonsei Med J.
2009 Aug;50(4):455-463. 10.3349/ymj.2009.50.4.455.
Epigenetic Cross-Talk between DNA Methylation and Histone Modifications in Human Cancers
- Affiliations
-
- 1Division of Molecular Oncology, Aichi Cancer Center Research Institute, Nagoya, Japan. ykondo@aichi-cc.jp
- KMID: 1758603
- DOI: http://doi.org/10.3349/ymj.2009.50.4.455
Abstract
- DNA methylation, histone modifications, and the chromatin structure are profoundly altered in human cancers. The silencing of cancer-related genes by these epigenetic regulators is recognized as a key mechanism in tumor formation. Recent findings revealed that DNA methylation and histone modifications appear to be linked to each other. However, it is not clearly understood how the formation of histone modifications may affect DNA methylation and which genes are relevantly involved with tumor formation. The presence of histone modifications does not always link to DNA methylation in human cancers, which suggests that another factor is required to connect these two epigenetic mechanisms. In this review, examples of studies that demonstrated the relationship between histone modifications and DNA methylation in human cancers are presented and the potential implications of these epigenetic mechanisms in human neoplasia are discussed.
MeSH Terms
Figure
Reference
-
1. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002. 3:415–428.
Article2. Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004. 23:29–39.
Article3. Turker MS. The establishment and maintenance of DNA methylation patterns in mouse somatic cells. Semin Cancer Biol. 1999. 9:329–337.
Article4. Bestor TH, Gundersen G, Kolstø AB, Prydz H. CpG islands in mammalian gene promoters are inherently resistant to de novo methylation. Genet Anal Tech Appl. 1992. 9:48–53.
Article5. Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995. 5:309–314.
Article6. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006. 6:107–116.
Article7. Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999. 98:285–294.
Article8. Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000. 403:41–45.9. Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999. 99:451–454.
Article10. Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000. 103:263–271.
Article11. Jenuwein T, Allis CD. Translating the histone code. Science. 2001. 293:1074–1080.
Article12. Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci U S A. 2004. 101:7398–7403.
Article13. Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006. 8:188–194.14. Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007. 39:237–242.
Article15. Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007. 39:157–158.
Article16. Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007. 39:232–236.
Article17. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002. 298:1039–1043.
Article18. Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008. 40:741–750.
Article19. Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008. 105:12979–12984.
Article20. Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998. 8:140–146.
Article21. Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006. 125:213–217.
Article22. Roth SY, Allis CD. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996. 87:5–8.
Article23. Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004. 14:155–164.
Article24. Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002. 22:6070–6078.
Article25. Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999. 98:37–46.
Article26. Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V. Polycomb silencing blocks transcription initiation. Mol Cell. 2004. 13:887–893.
Article27. Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006. 125:315–326.
Article28. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007. 448:553–560.
Article29. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007. 128:683–692.
Article30. Cadieux B, Ching TT, VandenBerg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006. 66:8469–8476.
Article31. Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995. 55:4525–4530.32. Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998. 18:6457–6473.
Article33. Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997. 57:808–811.34. Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994. 269:17228–17237.
Article35. Toyota M, Shen L, Ohe-Toyota M, Hamilton SR, Sinicrope FA, Issa JP. Aberrant methylation of the Cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res. 2000. 60:4044–4048.36. Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999. 59:6087–6090.37. Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007. 448:714–717.
Article38. Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007. 27:2746–2757.
Article39. Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007. 39:457–466.
Article40. Godmann M, Auger V, Ferraroni-Aguiar V, Di Sauro A, Sette C, Behr R, et al. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol Reprod. 2007. 77:754–764.
Article41. Hotz HR, Peters AH. Protein demethylation required for DNA methylation. Nat Genet. 2009. 41:10–11.
Article42. Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007. 131:633–636.
Article43. Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007. 449:248–251.
Article44. Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007. 130:77–88.
Article45. Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007. 450:908–912.46. Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009. 41:125–129.
Article47. Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007. 448:318–324.
Article48. Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008. 454:49–55.
Article49. Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998. 393:386–389.
Article50. Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998. 18:6538–6547.
Article51. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002. 16:6–21.
Article52. Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001. 414:277–283.
Article53. Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008. 15:1176–1183.
Article54. Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008. 27:2691–2701.
Article55. Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008. 27:2681–2690.
Article56. McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009. 69:282–291.
Article57. Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006. 439:871–874.
Article58. Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008. 2:160–169.
Article59. Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006. 5:37–50.
Article60. van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999. 23:474–478.
Article61. Issa JP. Decitabine. Curr Opin Oncol. 2003. 15:446–451.
Article62. McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006. 66:3541–3549.
Article63. Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999. 21:103–107.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Aberrant Epigenetic Modifications of LPHN2 Function as a Potential Cisplatin-Specific Biomarker for Human Gastrointestinal Cancer
- Epigenetic modifications and diabetic nephropathy
- Writing, erasing and reading histone lysine methylations
- Epigenetic regulation and chromatin remodeling in learning and memory
- The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis