Cancer Res Treat.
2019 Jan;51(1):391-401. 10.4143/crt.2018.103.
NFATC3–PLA2G15 Fusion Transcript Identified by RNA Sequencing Promotes Tumor Invasion and Proliferation in Colorectal Cancer Cell Lines
- Affiliations
-
- 1Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea. saewon1@snu.ac.kr kimty@snu.ac.kr
- 2Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University College of Medicine, Seoul, Korea.
- 3Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea.
- 4Department of Chemistry, College of Science, Yonsei University, Seoul, Korea.
- KMID: 2437629
- DOI: http://doi.org/10.4143/crt.2018.103
Abstract
- PURPOSE
This study was designed to identify novel fusion transcripts (FTs) and their functional significance in colorectal cancer (CRC) lines.
MATERIALS AND METHODS
We performed paired-end RNA sequencing of 28 CRC cell lines. FT candidates were identified using TopHat-fusion, ChimeraScan, and FusionMap tools and further experimental validation was conducted through reverse transcription-polymerase chain reaction and Sanger sequencing. FT was depleted in human CRC line and the effects on cell proliferation, cell migration, and cell invasion were analyzed.
RESULTS
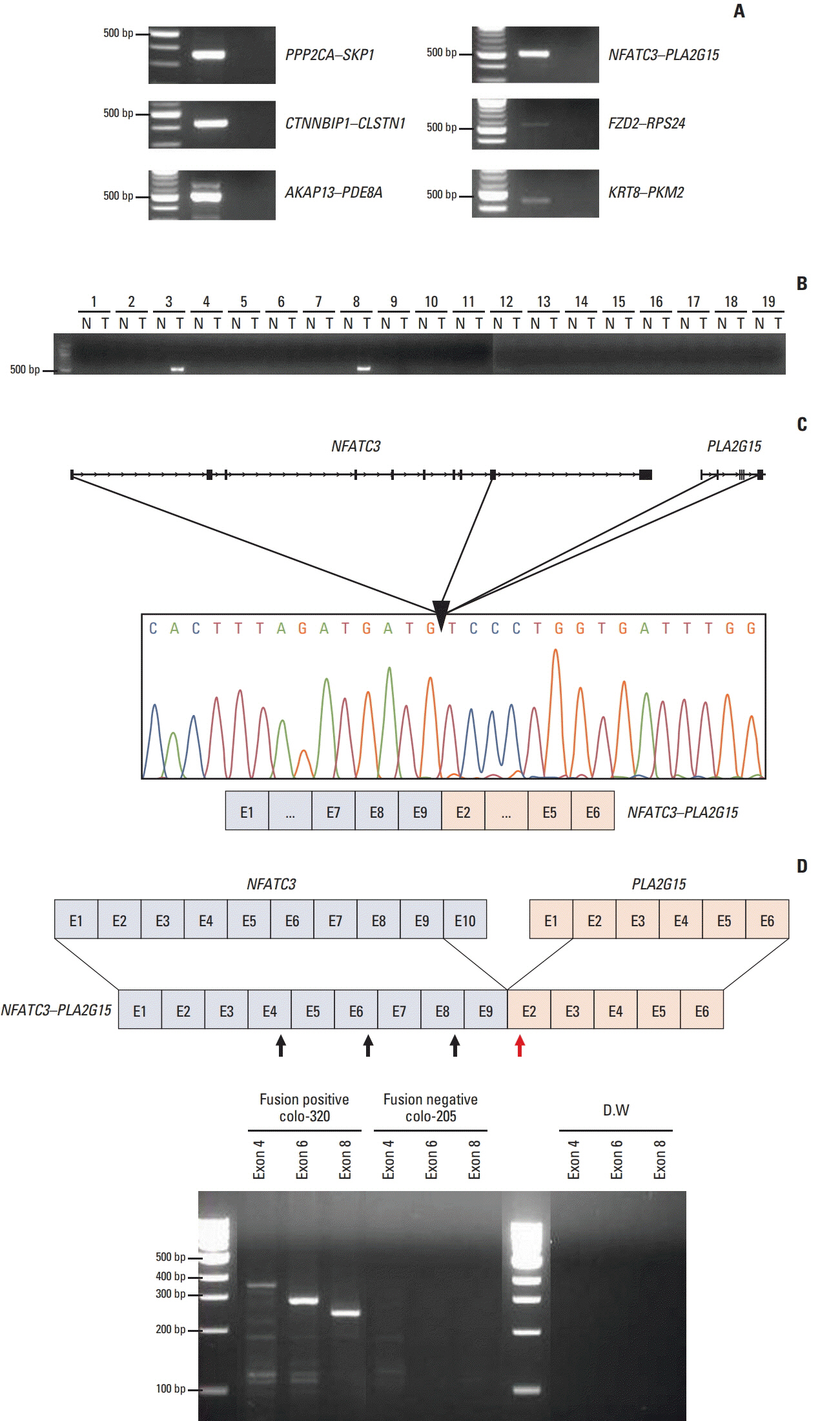
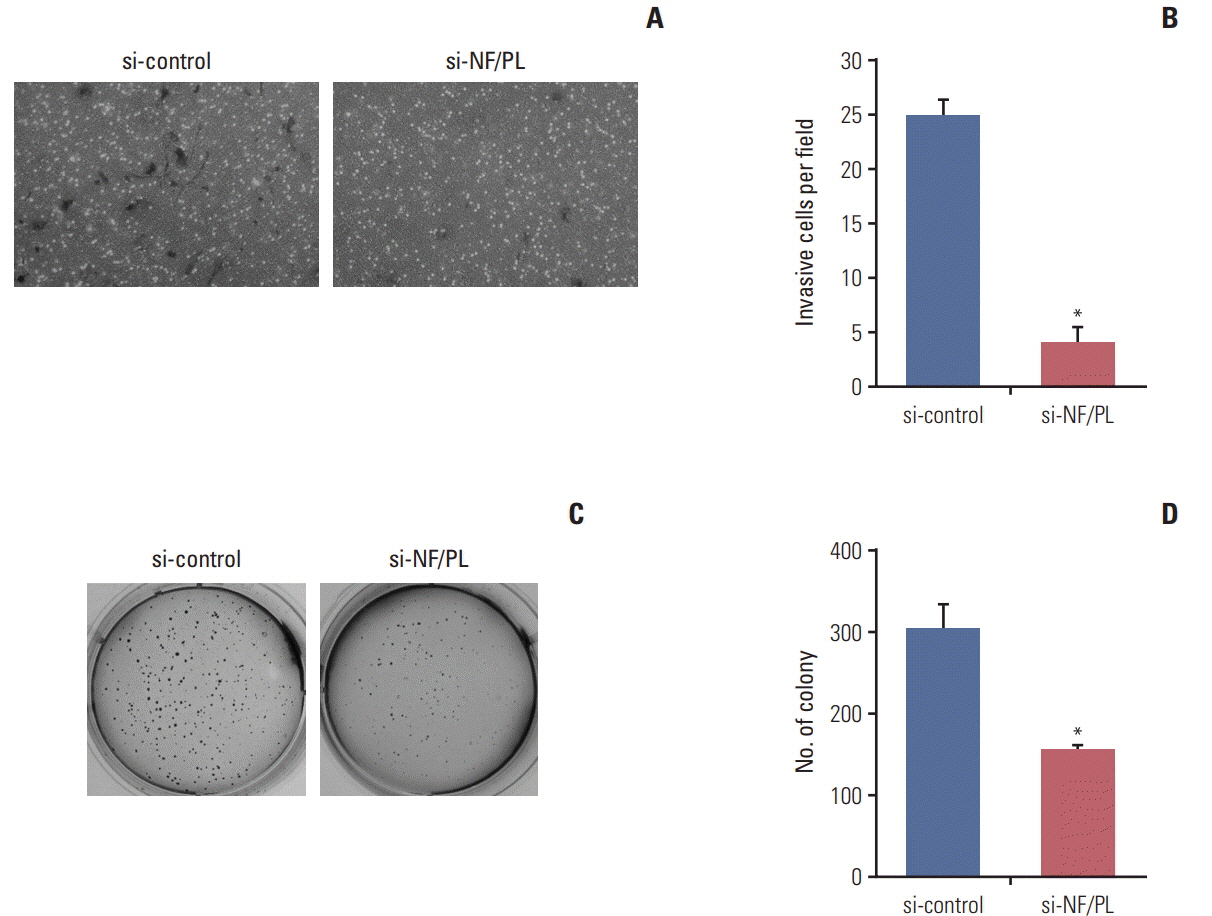
One thousand three hundred eighty FT candidates were detected through bioinformatics filtering. We selected six candidate FTs, including four inter-chromosomal and two intrachromosomal FTs and each FT was found in at least one of the 28 cell lines. Moreover, when we tested 19 pairs of CRC tumor and adjacent normal tissue samples, NFATC3-PLA2G15 FT was found in two. Knockdown of NFATC3-PLA2G15 using siRNA reduced mRNA expression of epithelial-mesenchymal transition (EMT) markers such as vimentin, twist, and fibronectin and increased mesenchymal-epithelial transition markers of E-cadherin, claudin-1, and FOXC2 in colo-320 cell line harboring NFATC3-PLA2G15 FT. The NFATC3-PLA2G15 knockdown also inhibited invasion, colony formation capacity, and cell proliferation.
CONCLUSION
These results suggest that that NFATC3-PLA2G15 FTs may contribute to tumor progression by enhancing invasion by EMT and proliferation.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014; 64:104–17.
Article2. Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002; 2:750–63.
Article3. Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009; 106:12353–8.
Article4. Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009; 458:97–101.
Article5. Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011; 12:87–98.
Article6. Kim M, Lee KH, Yoon SW, Kim BS, Chun J, Yi H. Analytical tools and databases for metagenomics in the next-generation sequencing era. Genomics Inform. 2013; 11:102–13.
Article7. Griswold IJ, MacPartlin M, Bumm T, Goss VL, O'Hare T, Lee KA, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol. 2006; 26:6082–93.
Article8. Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013; 13:443–54.
Article9. Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007; 7:233–45.
Article10. Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013; 13:772–87.
Article11. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007; 448:561–6.
Article12. Antoniu SA. Crizotinib for EML4-ALK positive lung adenocarcinoma: a hope for the advanced disease? Evaluation of Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363(18):1693-703. Expert Opin Ther Targets. 2011; 15:351–3.13. Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011; 43:964–8.
Article14. Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012; 488:660–4.
Article15. Nome T, Hoff AM, Bakken AC, Rognum TO, Nesbakken A, Skotheim RI. High frequency of fusion transcripts involving TCF7L2 in colorectal cancer: novel fusion partner and splice variants. PLoS One. 2014; 9:e91264.
Article16. Nome T, Thomassen GO, Bruun J, Ahlquist T, Bakken AC, Hoff AM, et al. Common fusion transcripts identified in colorectal cancer cell lines by high-throughput RNA sequencing. Transl Oncol. 2013; 6:546–53.
Article17. Lovf M, Nome T, Bruun J, Eknaes M, Bakken AC, Mpindi JP, et al. A novel transcript, VNN1-AB, as a biomarker for colorectal cancer. Int J Cancer. 2014; 135:2077–84.18. Ku JL, Park JG. Biology of SNU cell lines. Cancer Res Treat. 2005; 37:1–19.
Article19. Li X, Zhu L, Yang A, Lin J, Tang F, Jin S, et al. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell. 2011; 8:46–58.
Article20. Sengupta S, Jana S, Biswas S, Mandal PK, Bhattacharyya A. Cooperative involvement of NFAT and SnoN mediates transforming growth factor-beta (TGF-beta) induced EMT in metastatic breast cancer (MDA-MB 231) cells. Clin Exp Metastasis. 2013; 30:1019–31.21. Wen H, Li Y, Malek SN, Kim YC, Xu J, Chen P, et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS One. 2012; 7:e51203.
Article22. Bond J, Tran Quang C, Hypolite G, Belhocine M, Bergon A, Cordonnier G, et al. Novel intergenically spliced chimera, NFATC3-PLA2G15, is associated with aggressive T-ALL biology and outcome. Mol Cancer Res. 2018; 16:470–5.
Article23. Hoey T, Sun YL, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995; 2:461–72.
Article24. Oh-hora M, Rao A. The calcium/NFAT pathway: role in development and function of regulatory T cells. Microbes Infect. 2009; 11:612–9.
Article25. Branden LJ, Mohamed AJ, Smith CI. A peptide nucleic acidnuclear localization signal fusion that mediates nuclear transport of DNA. Nat Biotechnol. 1999; 17:784–7.
Article26. MacDonnell SM, Weisser-Thomas J, Kubo H, Hanscome M, Liu Q, Jaleel N, et al. CaMKII negatively regulates calcineurin-NFAT signaling in cardiac myocytes. Circ Res. 2009; 105:316–25.
Article27. Abe A, Kelly R, Shayman JA. The measurement of lysosomal phospholipase A2 activity in plasma. J Lipid Res. 2010; 51:2464–70.
Article28. Park JB, Lee CS, Jang JH, Ghim J, Kim YJ, You S, et al. Phospholipase signalling networks in cancer. Nat Rev Cancer. 2012; 12:782–92.
Article29. Nilsson LM, Sun ZW, Nilsson J, Nordstrom I, Chen YW, Molkentin JD, et al. Novel blocker of NFAT activation inhibits IL-6 production in human myometrial arteries and reduces vascular smooth muscle cell proliferation. Am J Physiol Cell Physiol. 2007; 292:C1167–78.
Article30. Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015; 6:7002.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Long Noncoding RNA HEIH Promotes Colorectal Cancer Tumorigenesis via Counteracting miR-939-Mediated Transcriptional Repression of Bcl-xL
- Identification of Alternative Splicing and Fusion Transcripts in Non-Small Cell Lung Cancer by RNA Sequencing
- The Role of CTGF in Osteosarcoma Progression
- Suppression of Antimicrobial Defense and Stabilization of STAT3 by IRAK-M Expression in Tumor Cells Promotes Colorectal Carcinogenesis
- CDK1 promotes the phosphorylation of KIFC1 to regulate the tumorgenicity of endometrial carcinoma