Tuberc Respir Dis.
2016 Apr;79(2):85-90. 10.4046/trd.2016.79.2.85.
Identification of Alternative Splicing and Fusion Transcripts in Non-Small Cell Lung Cancer by RNA Sequencing
- Affiliations
-
- 1Department of Internal Medicine, Kangwon National University School of Medicine, Chuncheon, Korea. pulmo2@kangwon.ac.kr
- 2Department of Oncology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- 3Department of Pulmonary and Critical Care Medicine and Clinical Research Center for Chronic Obstructive Airway Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2320770
- DOI: http://doi.org/10.4046/trd.2016.79.2.85
Abstract
- BACKGROUND
Lung cancer is the most common cause of cancer related death. Alterations in gene sequence, structure, and expression have an important role in the pathogenesis of lung cancer. Fusion genes and alternative splicing of cancer-related genes have the potential to be oncogenic. In the current study, we performed RNA-sequencing (RNA-seq) to investigate potential fusion genes and alternative splicing in non-small cell lung cancer.
METHODS
RNA was isolated from lung tissues obtained from 86 subjects with lung cancer. The RNA samples from lung cancer and normal tissues were processed with RNA-seq using the HiSeq 2000 system. Fusion genes were evaluated using Defuse and ChimeraScan. Candidate fusion transcripts were validated by Sanger sequencing. Alternative splicing was analyzed using multivariate analysis of transcript sequencing and validated using quantitative real time polymerase chain reaction.
RESULTS
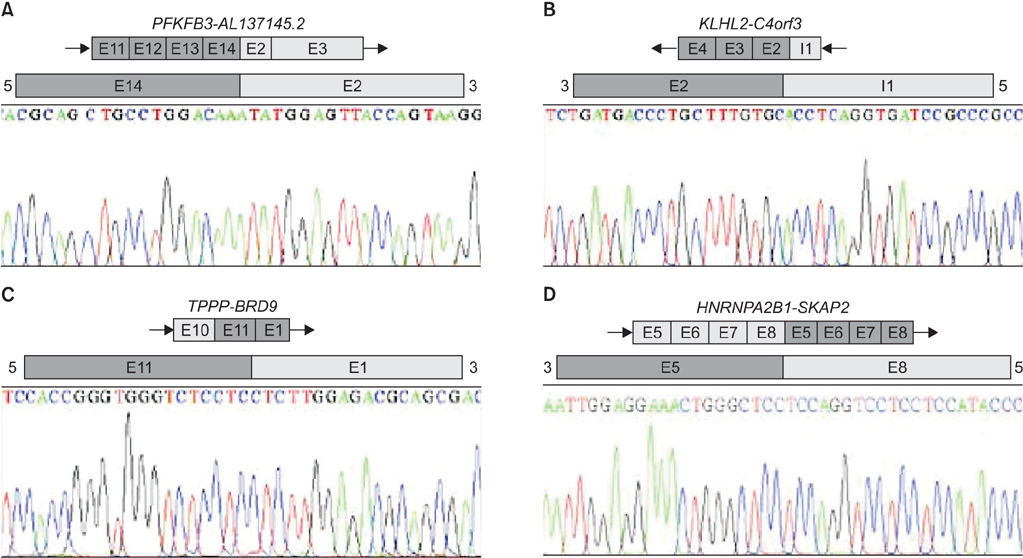
RNA-seq data identified oncogenic fusion genes EML4-ALK and SLC34A2-ROS1 in three of 86 normal-cancer paired samples. Nine distinct fusion transcripts were selected using DeFuse and ChimeraScan; of which, four fusion transcripts were validated by Sanger sequencing. In 33 squamous cell carcinoma, 29 tumor specific skipped exon events and six mutually exclusive exon events were identified. ITGB4 and PYCR1 were top genes that showed significant tumor specific splice variants.
CONCLUSION
In conclusion, RNA-seq data identified novel potential fusion transcripts and splice variants. Further evaluation of their functional significance in the pathogenesis of lung cancer is required.
MeSH Terms
Figure
Cited by 1 articles
-
Plasma CRABP2 as a Novel Biomarker in Patients with Non-Small Cell Lung Cancer
Do Jun Kim, Woo Jin Kim, Myoungnam Lim, Yoonki Hong, Seung-Joon Lee, Seok-Ho Hong, Jeongwon Heo, Hui-Young Lee, Seon-Sook Han
J Korean Med Sci. 2018;33(26):. doi: 10.3346/jkms.2018.33.e178.
Reference
-
1. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007; 448:561–566.2. Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012; 22:2109–2119.3. Kim SC, Jung Y, Park J, Cho S, Seo C, Kim J, et al. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS One. 2013; 8:e55596.4. Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011; 12:671–682.5. Kim YJ, Kim HS. Alternative splicing and its impact as a cancer diagnostic marker. Genomics Inform. 2012; 10:74–80.6. Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003; 72:291–336.7. Feng H, Qin Z, Zhang X. Opportunities and methods for studying alternative splicing in cancer with RNA-Seq. Cancer Lett. 2013; 340:179–191.8. Miura K, Fujibuchi W, Unno M. Splice isoforms as therapeutic targets for colorectal cancer. Carcinogenesis. 2012; 33:2311–2319.9. McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011; 7:e1001138.10. Iyer MK, Chinnaiyan AM, Maher CA. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011; 27:2903–2904.11. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111.12. Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012; 40:e61.13. Misquitta-Ali CM, Cheng E, O'Hanlon D, Liu N, McGlade CJ, Tsao MS, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. 2011; 31:138–150.14. Bando H, Atsumi T, Nishio T, Niwa H, Mishima S, Shimizu C, et al. Phosphorylation of the 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatase/PFKFB3 family of glycolytic regulators in human cancer. Clin Cancer Res. 2005; 11:5784–5792.15. Clem BF, O'Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA 2nd, et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013; 12:1461–1470.16. Dhanoa BS, Cogliati T, Satish AG, Bruford EA, Friedman JS. Update on the Kelch-like (KLHL) gene family. Hum Genomics. 2013; 7:13.17. Alsarraj J, Hunter KW. Bromodomain-containing protein 4: a dynamic regulator of breast cancer metastasis through modulation of the extracellular matrix. Int J Breast Cancer. 2012; 2012:670632.18. Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakacs A, Coppola L, et al. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011; 71:4464–4472.19. Shimamura S, Sasaki K, Tanaka M. The Src substrate SKAP2 regulates actin assembly by interacting with WAVE2 and cortactin proteins. J Biol Chem. 2013; 288:1171–1183.20. Banky B, Raso-Barnett L, Barbai T, Timar J, Becsagh P, Raso E. Characteristics of CD44 alternative splice pattern in the course of human colorectal adenocarcinoma progression. Mol Cancer. 2012; 11:83.21. Zygalaki E, Tsaroucha EG, Kaklamanis L, Lianidou ES. Quantitative real-time reverse transcription PCR study of the expression of vascular endothelial growth factor (VEGF) splice variants and VEGF receptors (VEGFR-1 and VEGFR-2) in non small cell lung cancer. Clin Chem. 2007; 53:1433–1439.22. Giancotti FG. Targeting integrin beta4 for cancer and antiangiogenic therapy. Trends Pharmacol Sci. 2007; 28:506–511.23. Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006; 7:325.24. Jariwala U, Prescott J, Jia L, Barski A, Pregizer S, Cogan JP, et al. Identification of novel androgen receptor target genes in prostate cancer. Mol Cancer. 2007; 6:39.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of HuD (a paraneoplastic encephalomyelitis antigen) mRNA in lung cancer
- Identification of neoantigens derived from alternative splicing and RNA modification
- Alternative Splicing and Its Impact as a Cancer Diagnostic Marker
- FusionScan: accurate prediction of fusion genes from RNA-Seq data
- In Vivo Target RNA Specificity of Trans-Splicing Phenomena by the Group I Intron