J Vet Sci.
2018 Nov;19(6):771-781. 10.4142/jvs.2018.19.6.771.
Molecular prophage typing of Staphylococcus aureus isolates from bovine mastitis
- Affiliations
-
- 1Department of Farm Animal Medicine, College of Veterinary Medicine and BK21 for Veterinary Science, Seoul National University, Seoul 08826, Korea. kwonhj01@snu.ac.kr
- 2Laboratory of Avian Diseases, College of Veterinary Medicine and BK21 for Veterinary Science, Seoul National University, Seoul 08826, Korea.
- 3Research Institute for Veterinary Science, College of Veterinary Medicine and BK21 for Veterinary Science, Seoul National University, Seoul 08826, Korea.
- KMID: 2427025
- DOI: http://doi.org/10.4142/jvs.2018.19.6.771
Abstract
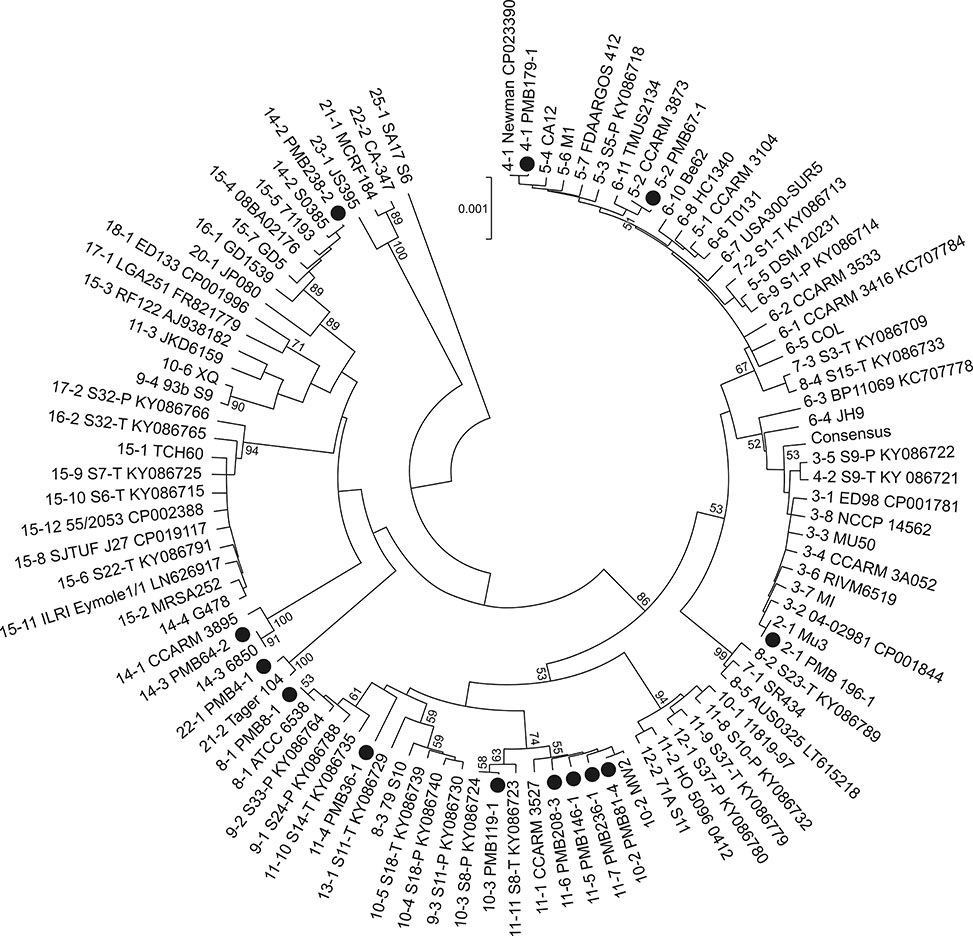
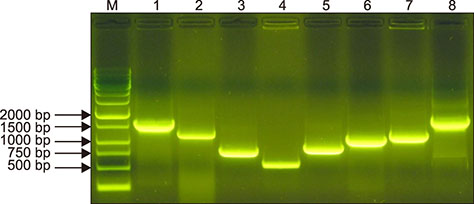
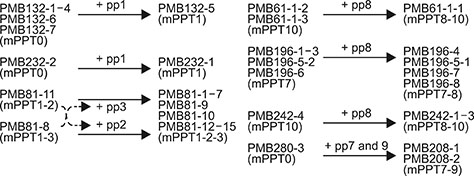
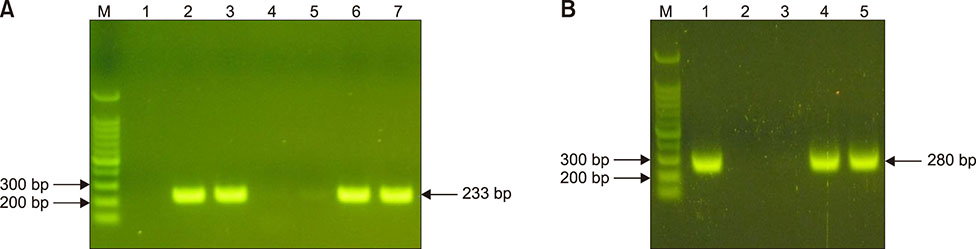
- Staphylococcus aureus is one of the major pathogens causing bovine mastitis and foodborne diseases associated with dairy products. To determine the genetic relationships between human and bovine or bovine isolates of S. aureus, various molecular methods have been used. Previously we developed an rpoB sequence typing (RSTing) method for molecular differentiation of S. aureus isolates and identification of RpoB-related antibiotic resistance. In this study, we performed spa typing and RSTing with 84 isolates from mastitic cows (22 farms, 72 cows, and 84 udders) and developed a molecular prophage typing (mPPTing) method for molecular epidemiological analysis of bovine mastitis. To compare the results, human isolates from patients (n = 14) and GenBank (n = 166) were used for real and in silico RSTing and mPPTing, respectively. Based on the results, RST10-2 and RST4-1 were the most common rpoB sequence types (RSTs) in cows and humans, respectively, and most isolates from cows and humans clearly differed. Antibiotic resistance-related RSTs were not detected in the cow isolates. A single dominant prophage type and gradual evolution through prophage acquisition were apparent in most of the tested farms. Thus, RSTing and mPPTing are informative, simple, and economic methods for molecular epidemiological analysis of S. aureus infections.
Keyword
MeSH Terms
Figure
Reference
-
1. Aubry-Damon H, Soussy CJ, Courvalin P. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1998; 42:2590–2594.
Article2. Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006; 62:1035–1047.
Article3. Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989; 43:267–292.
Article4. Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003; 49:277–300.
Article5. Cassat JE, Dunman PM, McAleese F, Murphy E, Projan SJ, Smeltzer MS. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J Bacteriol. 2005; 187:576–592.
Article6. Cui L, Isii T, Fukuda M, Ochiai T, Neoh HM, Camargo IL, Watanabe Y, Shoji M, Hishinuma T, Hiramatsu K. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2010; 54:5222–5233.
Article7. Devriese LA, Oeding P. Characteristics of Staphylococcus aureus strains isolated from different animal species. Res Vet Sci. 1976; 21:284–291.
Article8. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000; 38:1008–1015.
Article9. Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009; 191:3462–3468.
Article10. Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001; 39:4190–4192.
Article11. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003; 41:5442–5448.
Article12. Huijsdens XW, van Dijke BJ, Spalburg E, van Santen-Verheuvel MG, Heck ME, Pluister GN, Voss A, Wannet WJ, de Neeling AJ. Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob. 2006; 5:26.13. Hwang SY, Park YK, Koo HC, Park YH. spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea. J Vet Sci. 2010; 11:125–131.
Article14. Iandolo JJ, Worrell V, Groicher KH, Qian Y, Tian R, Kenton S, Dorman A, Ji H, Lin S, Loh P, Qi S, Zhu H, Roe BA. Comparative analysis of the genomes of the temperate bacteriophages φ11, φ12 and φ13 of Staphylococcus aureus 8325. Gene. 2002; 289:109–118.
Article15. Jevons MP. “Celbenin” - resistant Staphylococci. Br Med J. 1961; 1:124–125.
Article16. Juhász-Kaszanyitzky E, Jánosi S, Somogyi P, Dán A, van der Graaf-van Bloois L, van Duijkeren E, Wagenaar JA. MRSA transmission between cows and humans. Emerg Infect Dis. 2007; 13:630–632.
Article17. Kapur V, Sischo WM, Greer RS, Whittam TS, Musser JM. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J Clin Microbiol. 1995; 33:376–380.
Article18. Kim D, Kim EK, Seong WJ, Ro Y, Ko DS, Kim NH, Kim JH, Kwon HJ. Identification of microbiome with 16S rRNA gene pyrosequencing and antimicrobial effect of egg white in bovine mastitis. Korean J Vet Res. 2017; 57:117–126. Korean.
Article19. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007; 51:264–274.
Article20. Kraushaar B, Hammerl JA, Kienöl M, Heinig ML, Sperling N, Dinh Thanh M, Reetz J, Jäckel C, Fetsch A, Hertwig S. Acquisition of virulence factors in livestock-associated MRSA: lysogenic conversion of CC398 strains by virulence gene-containing phages. Sci Rep. 2017; 7:2004.
Article21. Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983; 305:709–712.
Article22. Kuehn BM. Antibiotic-resistant "superbugs" may be transmitted from animals to humans. JAMA. 2007; 298:2125–2126.
Article23. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874.
Article24. Kwon HJ, Seong WJ, Kim JH. Molecular prophage typing of avian pathogenic Escherichia coli. Vet Microbiol. 2013; 162:785–792.25. Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res. 2003; 2:63–76.26. Lee JH. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003; 69:6489–6494.
Article27. Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, Miedzobrodzki J, Kosowska-Shick K, Appelbaum PC, Hryniewicz W. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J Clin Microbiol. 2005; 43:3095–3100.
Article28. Monecke S, Ruppelt A, Wendlandt S, Schwarz S, Slickers P, Ehricht R, Jäckel SC. Genotyping of Staphylococcus aureus isolates from diseased poultry. Vet Microbiol. 2013; 162:806–812.29. Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003; 49:93–105.
Article30. Resch G, François P, Morisset D, Stojanov M, Bonetti EJ, Schrenzel J, Sakwinska O, Moreillon P. Human-to-bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β-hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS One. 2013; 8:e58187.31. Sakwinska O, Giddey M, Moreillon M, Morisset D, Waldvogel A, Moreillon P. Staphylococcus aureus host range and human-bovine host shift. Appl Environ Microbiol. 2011; 77:5908–5915.
Article32. Schmidt T, Kock MM, Ehlers MM. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: genetic diversity and inter-species host transmission. Front Microbiol. 2017; 8:511.
Article33. Seong WJ, Kim JH, Kwon HJ. Comparison of complete rpoB gene sequence typing and multi-locus sequence typing for phylogenetic analysis of Staphylococcus aureus. J Gen Appl Microbiol. 2013; 59:335–343.
Article34. Seong WJ, Kwon HJ, Kim TE, Lee DY, Park MS, Kim JH. Molecular serotyping of Salmonella enterica by complete rpoB gene sequencing. J Microbiol. 2012; 50:962–969.
Article35. Serwer P, Hayes SJ, Zaman S, Lieman K, Rolando M, Hardies SC. Improved isolation of undersampled bacteriophages: finding of distant terminase genes. Virology. 2004; 329:412–424.
Article36. Silva NC, Guimarães FF, Manzi MP, Budri PE, Gómez-Sanz E, Benito D, Langoni H, Rall VL, Torres C. Molecular characterization and clonal diversity of methicillin-susceptible Staphylococcus aureus in milk of cows with mastitis in Brazil. J Dairy Sci. 2013; 96:6856–6862.
Article37. Spoor LE, McAdam PR, Weinert LA, Rambaut A, Hasman H, Aarestrup FM, Kearns AM, Larsen AR, Skov RL, Fitzgerald JR. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. MBio. 2013; 4:e00356–e00313.
Article38. Strommenger B, Braulke C, Heuck D, Schmidt C, Pasemann B, Nübel U, Witte W. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008; 46:574–581.
Article39. Sung JM, Lloyd DH, Lindsay JA. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008; 154:1949–1959.
Article40. Sutra L, Poutrel B. Virulence factors involved in the pathogenesis of bovine intramammary infections due to Staphylococcus aureus. J Med Microbiol. 1994; 40:79–89.
Article41. Suzuki M, Matsumoto M, Takahashi M, Hayakawa Y, Minagawa H. Identification of the clonal complexes of Staphylococcus aureus strains by determination of the conservation patterns of small genomic islets. J Appl Microbiol. 2009; 107:1367–1374.
Article42. Thompson RL, Cabezudo I, Wenzel RP. Epidemiology of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. Ann Intern Med. 1982; 97:309–317.
Article43. van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Voss A, Kluytmans J. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007; 13:1834–1839.
Article44. van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J Bacteriol. 2006; 188:1310–1315.
Article45. Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol. 2011; 49:2680–2684.
Article46. Wichelhaus TA, Schäfer V, Brade V, Böddinghaus B. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999; 43:2813–2816.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea
- Capsular polysaccharide typing of domestic mastitis-causing Staphylococcus aureus strains and its potential exploration of bovine mastitis vaccine developmen. I. capsular polysaccharide typing, isolation and purification of the strains
- Invasive potential of biofilm-forming Staphylococci bovine subclinical mastitis isolates
- Comparative studies on pheno- and genotypic properties of Staphylococcus aureus isolated from bovine subclinical mastitis in central Java in Indonesia and Hesse in Germany
- PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from bovine subclinical mastitis cases