Korean J Physiol Pharmacol.
2018 Jul;22(4):457-465. 10.4196/kjpp.2018.22.4.457.
Effect of BIS depletion on HSF1-dependent transcriptional activation in A549 non-small cell lung cancer cells
- Affiliations
-
- 1Department of Biochemistry, College of Medicine, The Catholic University of Korea, Seoul 06591, Korea. leejh@catholic.ac.kr
- 2The Institute for Aging and Metabolic Diseases, College of Medicine, The Catholic University of Korea, Seoul 06591, Korea.
- 3Laboratory of Genomic Instability and Cancer Therapeutics, Cancer Mutation Research Center, Chosun University School of medicine, Gwangju 61452, Korea.
- 4Genome Editing Research Center, KRIBB, Daejeon 34141, Korea.
- 5Department of Biomolecular Science, Korea University of Science and Technology, Daejeon 34113, Korea.
- KMID: 2414270
- DOI: http://doi.org/10.4196/kjpp.2018.22.4.457
Abstract
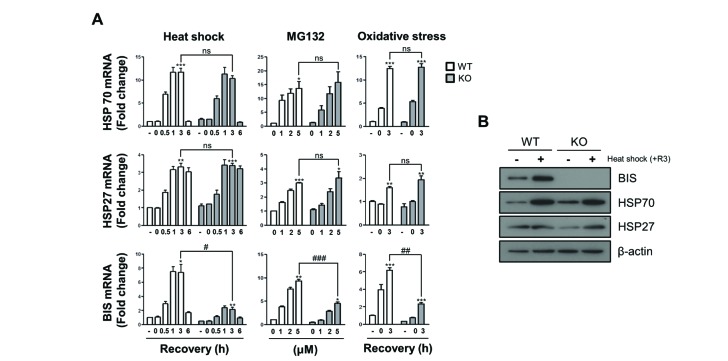
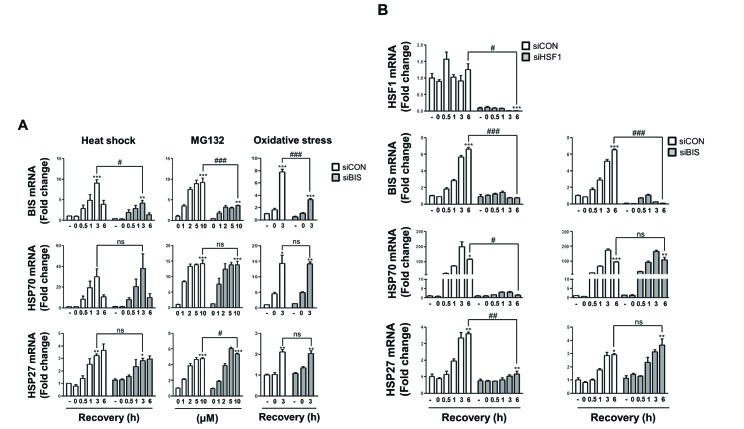
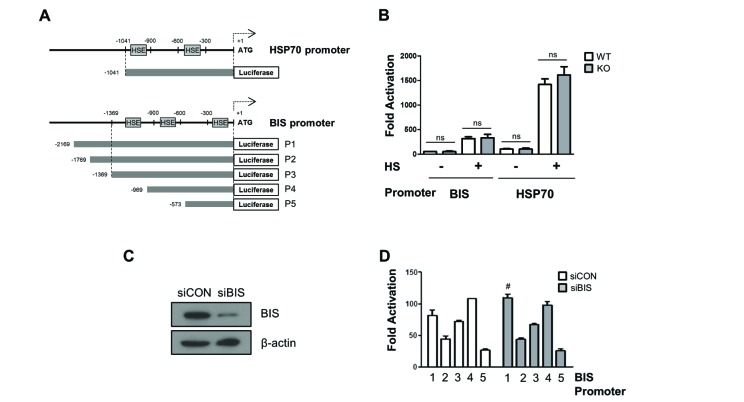
- The expression of BCL-2 interacting cell death suppressor (BIS), an anti-stress or anti-apoptotic protein, has been shown to be regulated at the transcriptional level by heat shock factor 1 (HSF1) upon various stresses. Recently, HSF1 was also shown to bind to BIS, but the significance of these protein-protein interactions on HSF1 activity has not been fully defined. In the present study, we observed that complete depletion of BIS using a CRISPR/Cas9 system in A549 non-small cell lung cancer did not affect the induction of heat shock protein (HSP) 70 and HSP27 mRNAs under various stress conditions such as heat shock, proteotoxic stress, and oxidative stress. The lack of a functional association of BIS with HSF1 activity was also demonstrated by transient downregulation of BIS by siRNA in A549 and U87 glioblastoma cells. Endogenous BIS mRNA levels were significantly suppressed in BIS knockout (KO) A549 cells compared to BIS wild type (WT) A549 cells at the constitutive and inducible levels. The promoter activities of BIS and HSP70 as well as the degradation rate of BIS mRNA were not influenced by depletion of BIS. In addition, the expression levels of the mutant BIS construct, in which 14 bp were deleted as in BIS-KO A549 cells, were not different from those of the WT BIS construct, indicating that mRNA stability was not the mechanism for autoregulation of BIS. Our results suggested that BIS was not required for HSF1 activity, but was required for its own expression, which involved an HSF1-independent pathway.
MeSH Terms
Figure
Reference
-
1. Lee JH, Takahashi T, Yasuhara N, Inazawa J, Kamada S, Tsujimoto Y. Bis, a Bcl-2-binding protein that synergizes with Bcl-2 in preventing cell death. Oncogene. 1999; 18:6183–6190. PMID: 10597216.
Article2. Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999; 274:781–786. PMID: 9873016.
Article3. Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011; 2:e141. PMID: 21472004.
Article4. Rosati A, Ammirante M, Gentilella A, Basile A, Festa M, Pascale M, Marzullo L, Belisario MA, Tosco A, Franceschelli S, Moltedo O, Pagliuca G, Lerose R, Turco MC. Apoptosis inhibition in cancer cells: a novel molecular pathway that involves BAG3 protein. Int J Biochem Cell Biol. 2007; 39:1337–1342. PMID: 17493862.
Article5. Zhu H, Liu P, Li J. BAG3: a new therapeutic target of human cancers? Histol Histopathol. 2012; 27:257–261. PMID: 22237703.6. Selcen D, Muntoni F, Burton BK, Pegoraro E, Sewry C, Bite AV, Engel AG. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009; 65:83–89. PMID: 19085932.
Article7. Odgerel Z, Sarkozy A, Lee HS, McKenna C, Rankin J, Straub V, Lochmüller H, Paola F, D'Amico A, Bertini E, Bushby K, Goldfarb LG. Inheritance patterns and phenotypic features of myofibrillar myopathy associated with a BAG3 mutation. Neuromuscul Disord. 2010; 20:438–442. PMID: 20605452.
Article8. Lei Z, Brizzee C, Johnson GV. BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol Aging. 2015; 36:241–248. PMID: 25212465.
Article9. Seidel K, Vinet J, Dunnen WF, Brunt ER, Meister M, Boncoraglio A, Zijlstra MP, Boddeke HW, Rüb U, Kampinga HH, Carra S. The HSPB8-BAG3 chaperone complex is upregulated in astrocytes in the human brain affected by protein aggregation diseases. Neuropathol Appl Neurobiol. 2012; 38:39–53. PMID: 21696420.
Article10. Merabova N, Sariyer IK, Saribas AS, Knezevic T, Gordon J, Turco MC, Rosati A, Weaver M, Landry J, Khalili K. WW domain of BAG3 is required for the induction of autophagy in glioma cells. J Cell Physiol. 2015; 230:831–841. PMID: 25204229.
Article11. Behl C. Breaking BAG: the Co-Chaperone BAG3 in health and disease. Trends Pharmacol Sci. 2016; 37:672–688. PMID: 27162137.
Article12. Rosati A, Basile A, Falco A, d'Avenia M, Festa M, Graziano V, De Laurenzi V, Arra C, Pascale M, Turco MC. Role of BAG3 protein in leukemia cell survival and response to therapy. Biochim Biophys Acta. 2012; 1826:365–369. PMID: 22710027.
Article13. Wang HQ, Liu HM, Zhang HY, Guan Y, Du ZX. Transcriptional upregulation of BAG3 upon proteasome inhibition. Biochem Biophys Res Commun. 2008; 365:381–385. PMID: 17996194.
Article14. Rosati A, Leone A, Del Valle L, Amini S, Khalili K, Turco MC. Evidence for BAG3 modulation of HIV-1 gene transcription. J Cell Physiol. 2007; 210:676–683. PMID: 17187345.
Article15. Gentilella A, Khalili K. BAG3 expression is sustained by FGF2 in neural progenitor cells and impacts cell proliferation. Cell Cycle. 2010; 9:4245–4247. PMID: 20962586.
Article16. Liu J, Qu CB, Xue YX, Li Z, Wang P, Liu YH. MiR-143 enhances the antitumor activity of shikonin by targeting BAG3 expression in human glioblastoma stem cells. Biochem Biophys Res Commun. 2015; 468:105–112. PMID: 26541455.
Article17. Flum M, Kleemann M, Schneider H, Weis B, Fischer S, Handrick R, Otte K. miR-217-5p induces apoptosis by directly targeting PRKCI, BAG3, ITGAV and MAPK1 in colorectal cancer cells. J Cell Commun Signal. 2018; 12:451–466. PMID: 28905214.
Article18. d'Avenia M, Citro R, De Marco M, Veronese A, Rosati A, Visone R, Leptidis S, Philippen L, Vitale G, Cavallo A, Silverio A, Prota C, Gravina P, De Cola A, Carletti E, Coppola G, Gallo S, Provenza G, Bossone E, Piscione F, Hahne M, De Windt LJ, Turco MC, De Laurenzi V. A novel miR-371a-5p-mediated pathway, leading to BAG3 upregulation in cardiomyocytes in response to epinephrine, is lost in Takotsubo cardiomyopathy. Cell Death Dis. 2015; 6:e1948. PMID: 26512958.19. Ben Aicha S, Lessard J, Pelletier M, Fournier A, Calvo E, Labrie C. Transcriptional profiling of genes that are regulated by the endoplasmic reticulum-bound transcription factor AIbZIP/CREB3L4 in prostate cells. Physiol Genomics. 2007; 31:295–305. PMID: 17712038.20. Gentilella A, Passiatore G, Deshmane S, Turco MC, Khalili K. Activation of BAG3 by Egr-1 in response to FGF-2 in neuroblastoma cells. Oncogene. 2008; 27:5011–5018. PMID: 18469860.
Article21. Cesaro E, Montano G, Rosati A, Crescitelli R, Izzo P, Turco MC, Costanzo P. WT1 protein is a transcriptional activator of the antiapoptotic bag3 gene. Leukemia. 2010; 24:1204–1206. PMID: 20410921.
Article22. Song S, Kole S, Precht P, Pazin MJ, Bernier M. Activation of heat shock factor 1 plays a role in pyrrolidine dithiocarbamate-mediated expression of the co-chaperone BAG3. Int J Biochem Cell Biol. 2010; 42:1856–1863. PMID: 20692357.
Article23. Yoo HJ, Im CN, Youn DY, Yun HH, Lee JH. Bis is induced by oxidative stress via activation of HSF1. Korean J Physiol Pharmacol. 2014; 18:403–409. PMID: 25352760.24. Jacobs AT, Marnett LJ. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J Biol Chem. 2009; 284:9176–9183. PMID: 19179333.
Article25. Franceschelli S, Rosati A, Lerose R, De Nicola S, Turco MC, Pascale M. Bag3 gene expression is regulated by heat shock factor 1. J Cell Physiol. 2008; 215:575–577. PMID: 18286539.
Article26. Gentilella A, Khalili K. Autoregulation of co-chaperone BAG3 gene transcription. J Cell Biochem. 2009; 108:1117–1124. PMID: 19777443.
Article27. Gentilella A, Khalili K. BAG3 expression in glioblastoma cells promotes accumulation of ubiquitinated clients in an Hsp70-dependent manner. J Biol Chem. 2011; 286:9205–9215. PMID: 21233200.
Article28. Chen Y, Yang LN, Cheng L, Tu S, Guo SJ, Le HY, Xiong Q, Mo R, Li CY, Jeong JS, Jiang L, Blackshaw S, Bi LJ, Zhu H, Tao SC, Ge F. Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol Cell Proteomics. 2013; 12:2804–2819. PMID: 23824909.
Article29. Dai C, Sampson SB. HSF1: Guardian of proteostasis in cancer. Trends Cell Biol. 2016; 26:17–28. PMID: 26597576.
Article30. Dai C, Santagata S, Tang Z, Shi J, Cao J, Kwon H, Bronson RT, Whitesell L, Lindquist S. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J Clin Invest. 2012; 122:3742–3754. PMID: 22945628.
Article31. Cui MN, Yun HH, Lee NE, Kim HY, Im CN, Kim YS, Lee JH. Depletion of BIS sensitizes A549 cells to treatment with cisplatin. Mol Cell Toxicol. 2016; 12:63–71.
Article32. Baek JY, Yun HH, Im CN, Ko JH, Jeong SM, Lee JH. BIS overexpression does not affect the sensitivity of HEK 293T cells against apoptosis. Mol Cell Toxicol. 2017; 13:95–103.
Article33. Jelluma N, Yang X, Stokoe D, Evan GI, Dansen TB, Haas-Kogan DA. Glucose withdrawal induces oxidative stress followed by apoptosis in glioblastoma cells but not in normal human astrocytes. Mol Cancer Res. 2006; 4:319–330. PMID: 16687487.
Article34. Liu Y, Song XD, Liu W, Zhang TY, Zuo J. Glucose deprivation induces mitochondrial dysfunction and oxidative stress in PC12 cell line. J Cell Mol Med. 2003; 7:49–56. PMID: 12767261.
Article35. Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013; 1829:612–623. PMID: 23435113.
Article36. Karousis ED, Nasif S, Mühlemann O. Nonsense-mediated mRNA decay: novel mechanistic insights and biological impact. Wiley Interdiscip Rev RNA. 2016; 7:661–682. PMID: 27173476.
Article37. Kim HY, Kim YS, Yun HH, Im CN, Ko JH, Lee JH. ERK-mediated phosphorylation of BIS regulates nuclear translocation of HSF1 under oxidative stress. Exp Mol Med. 2016; 48:e260. PMID: 27659916.
Article38. Jin YH, Ahn SG, Kim SA. BAG3 affects the nucleocytoplasmic shuttling of HSF1 upon heat stress. Biochem Biophys Res Commun. 2015; 464:561–567. PMID: 26159920.
Article39. Im CN, Yun HH, Lee JH. Heat shock factor 1 depletion sensitizes A172 glioblastoma cells to temozolomide via suppression of cancer stem cell-like properties. Int J Mol Sci. 2017; 18:E468. PMID: 28241425.
Article40. Fang X, Bogomolovas J, Wu T, Zhang W, Liu C, Veevers J, Stroud MJ, Zhang Z, Ma X, Mu Y, Lao DH, Dalton ND, Gu Y, Wang C, Wang M, Liang Y, Lange S, Ouyang K, Peterson KL, Evans SM, Chen J. Loss-of-function mutations in co-chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest. 2017; 127:3189–3200. PMID: 28737513.
Article41. Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol. 2017; 27:895–905. PMID: 28890254.
Article42. Su KH, Dai C. Metabolic control of the proteotoxic stress response: implications in diabetes mellitus and neurodegenerative disorders. Cell Mol Life Sci. 2016; 73:4231–4248. PMID: 27289378.
Article43. Jiang S, Tu K, Fu Q, Schmitt DC, Zhou L, Lu N, Zhao Y. Multifaceted roles of HSF1 in cancer. Tumour Biol. 2015; 36:4923–4931. PMID: 26108999.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Bis is Induced by Oxidative Stress via Activation of HSF1
- Heme Oxygenase-1 via Transcriptional Activation of Nrf2 Attenuates Apoptosis in Lung Cancer Cells
- The Effect of Inhibition of Heme Oxygenase-1 on Chemosensitivity of Cisplatin in Lung Cancer Cells
- Antiproliferative effect of Citrus junos extracts on A549 human non-smallcell lung cancer cells
- Antimetastatic effect of fucoidan against non-small cell lung cancer by suppressing non-receptor tyrosine kinase and extracellular signalrelated kinase pathway