J Vet Sci.
2018 Mar;19(2):216-231. 10.4142/jvs.2018.19.2.216.
Analysis of protein expression in Brucella abortus mutants with different growth rates by two-dimensional gel electrophoresis and LC-MS/MS peptide analysis
- Affiliations
-
- 1Department of Infectious Diseases, College of Veterinary Medicine, Seoul National University, Seoul 08826, Korea. yoohs@snu.ac.kr
- 2Institute of Green-Bio Science and Technology, Seoul National University, Pyeongchang 25354, Korea.
- KMID: 2407621
- DOI: http://doi.org/10.4142/jvs.2018.19.2.216
Abstract
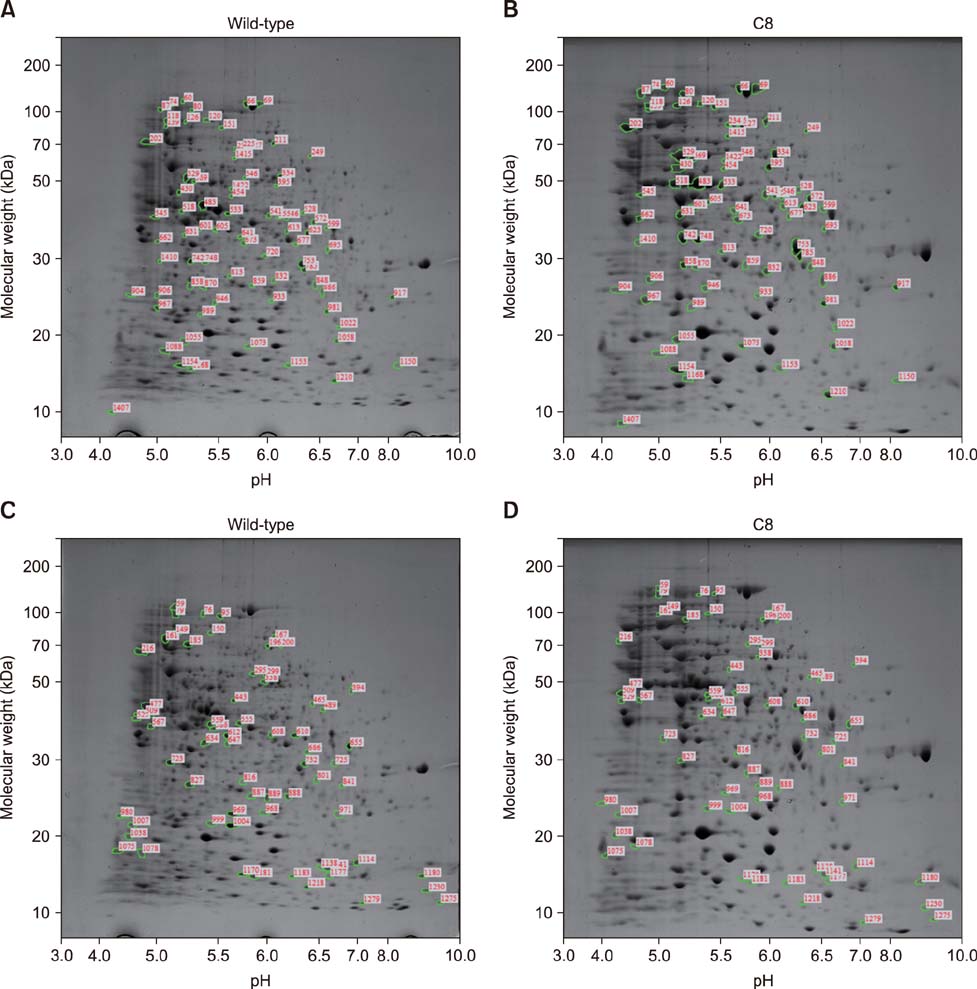
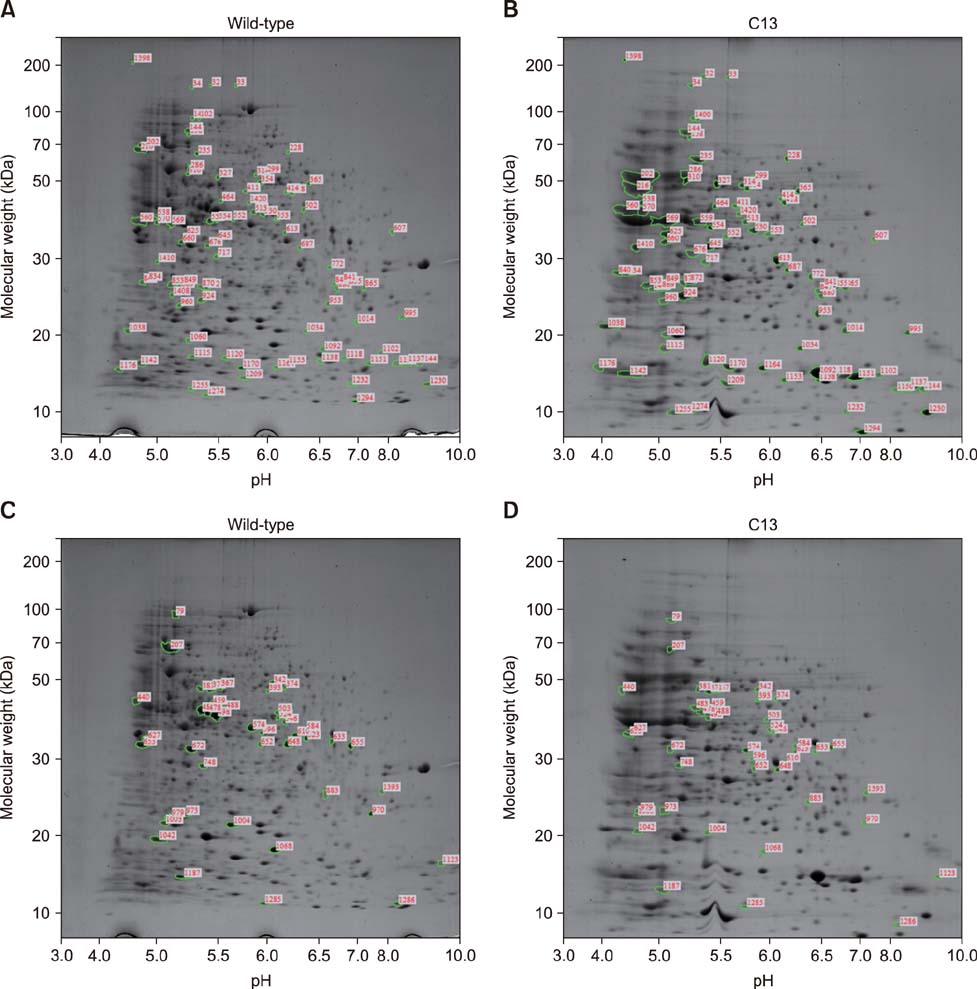
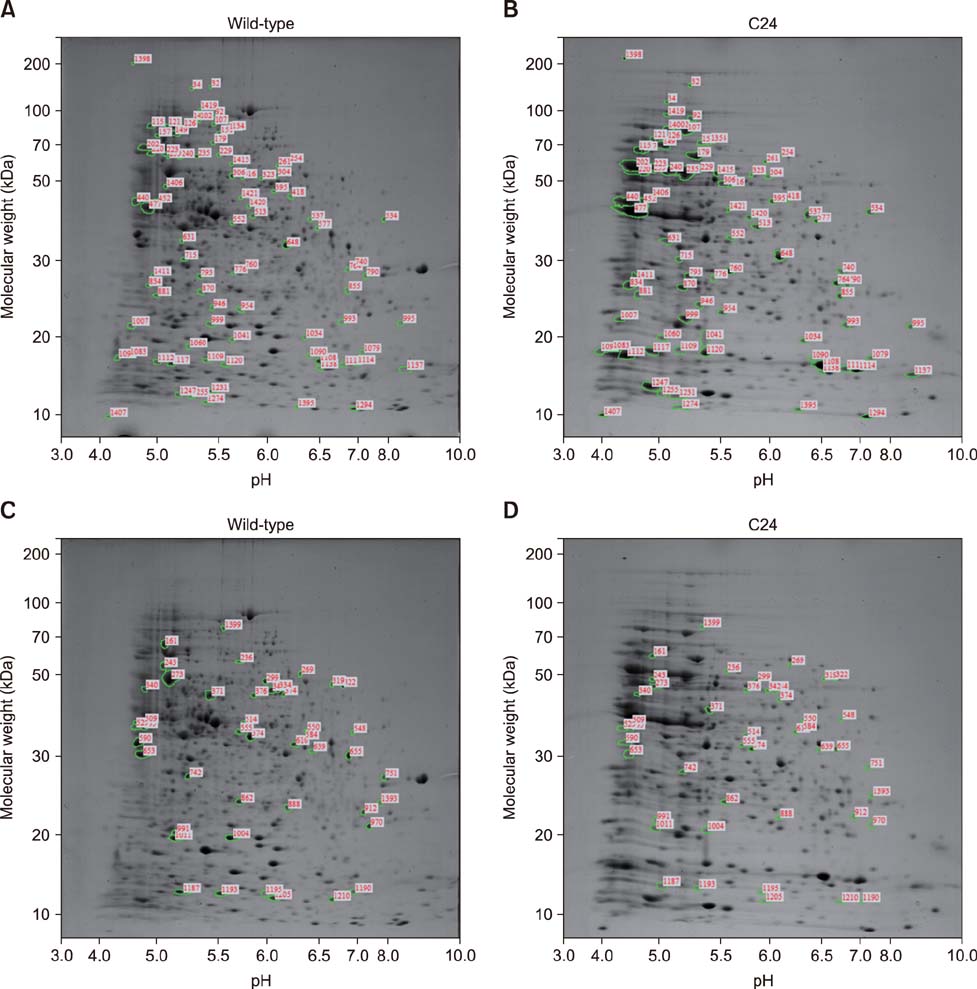
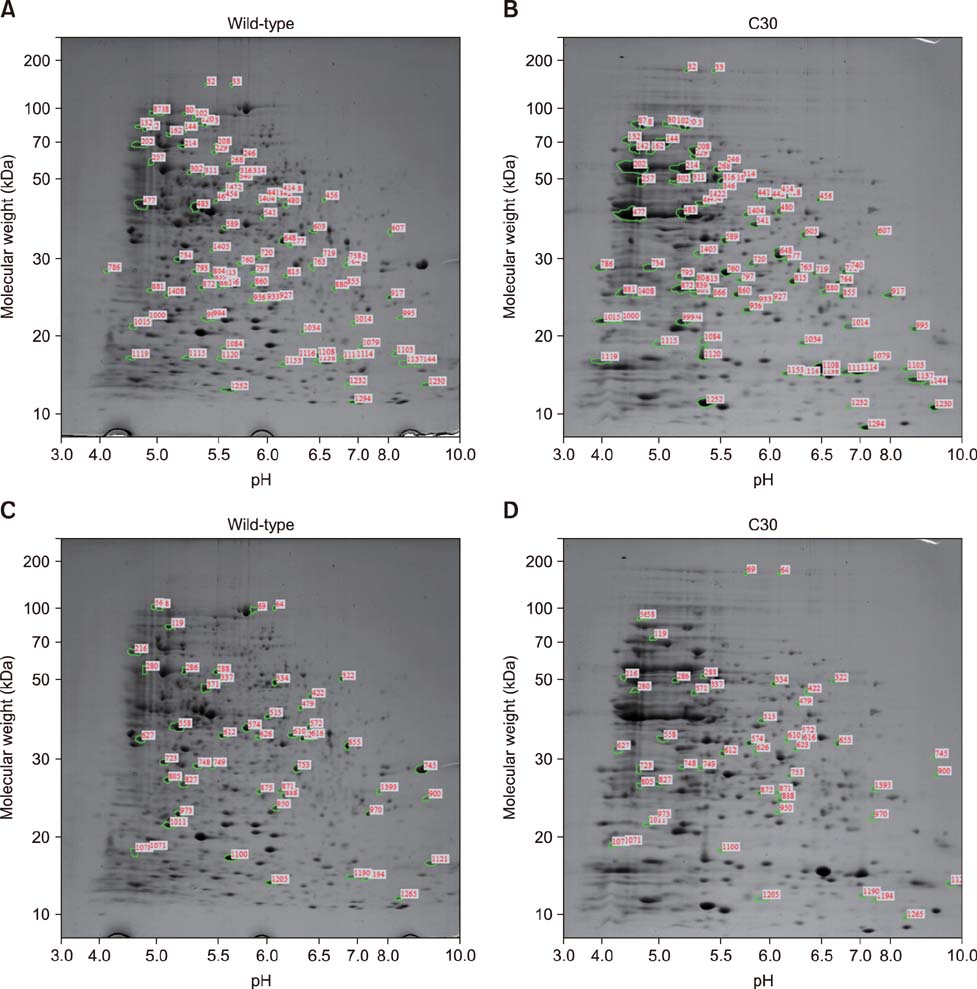
- Brucella abortus is a bacterium that causes brucellosis and is the causative agent of worldwide zoonoses. Pathogenesis of the B. abortus infection is complicated, and several researchers have attempted to elucidate the infection mechanism of B. abortus. While several proteins have been revealed as pathogenic factors by previous researchers, the underlying mechanism of B. abortus infection is unresolved. In this study, we identified proteins showing different expression levels in B. abortus mutants with different biological characteristics that were generated by random insertion of a transposon. Five mutants were selected based on biological characteristics, in particular, their growth features. Total proteins of mutant and wild-type B. abortus were purified and subjected to two-dimensional gel electrophoresis. Thirty protein spots of each mutant with expression increases or decreases were selected; those with a change of more than 2-fold were compared with the wild-type. Selected spots underwent liquid chromatography tandem mass spectrometry for peptide analysis. DnaK and ClpB, involved in protein aggregation, increased. SecA and GAPDH, associated with energy metabolism, decreased in some mutants with a growth rate slower than that of the wild-type. Mutants with slower growth showed a decrease in energy metabolism-related proteins, while mutants with faster growth showed an increase in pathogenicity-related proteins.
Keyword
MeSH Terms
Figure
Reference
-
1. Acebrón SP, Martín I, del Castillo U, Moro F, Muga A. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009; 583:2991–2996.
Article2. Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999; 21:932–939.
Article3. Boschiroli ML, Foulongne V, O'Callaghan D. Brucellosis: a worldwide zoonosis. Curr Opin Microbiol. 2001; 4:58–64.
Article4. Buchberger A, Theyssen H, Schröder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995; 270:16903–16910.
Article5. Cellier MF, Teyssier J, Nicolas M, Liautard JP, Marti J, Sri Widada J. Cloning and characterization of the Brucella ovis heat shock protein DnaK functionally expressed in Escherichia coli. J Bacteriol. 1992; 174:8036–8042.
Article6. Christopher S, Umapathy BL, Ravikumar KL. Brucellosis: review on the recent trends in pathogenicity and laboratory diagnosis. J Lab Physicians. 2010; 2:55–60.
Article7. Cui M, Wang T, Xu J, Ke Y, Du X, Yuan X, Wang Z, Gong C, Zhuang Y, Lei S, Su X, Wang X, Huang L, Zhong Z, Peng G, Yuan J, Chen Z, Wang Y. Impact of Hfq on global gene expression and intracellular survival in Brucella melitensis. PLoS One. 2013; 8:e71933.8. DelVecchio VG, Wagner MA, Eschenbrenner M, Horn TA, Kraycer JA, Estock F, Elzer P, Mujer CV. Brucella proteomes--a review. Vet Microbiol. 2002; 90:593–603.9. Driessen AJ. SecB, a molecular chaperone with two faces. Trends Microbiol. 2001; 9:193–196.
Article10. Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997; 61:136–169.
Article11. Franco MP, Mulder M, Gilman RH, Smits HL. Human brucellosis. Lancet Infect Dis. 2007; 7:775–786.
Article12. Gruer MJ, Guest JR. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology. 1994; 140:2531–2541.
Article13. Guzman-Verri C, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, Garin J, Gorvel JP, Moriyon I, Moreno E, Lopez-Goni I. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc Natl Acad Sci U S A. 2002; 99:12375–12380.14. Lamontagne J, Béland M, Forest A, Côté-Martin A, Nassif N, Tomaki F, Moriyón I, Moreno E, Paramithiotis E. Proteomics-based confirmation of protein expression and correction of annotation errors in the Brucella abortus genome. BMC Genomics. 2010; 11:300.15. Lee HJ, Cha HJ, Lim JS, Lee SH, Song SY, Kim H, Hancock WS, Yoo JS, Paik YK. Abundance-ratio-based semiquantitative analysis of site-specific N-linked glycopeptides present in the plasma of hepatocellular carcinoma patients. J Proteome Res. 2014; 13:2328–2338.
Article16. Lee J, Kim KY, Lee J, Paik YK. Regulation of Dauer formation by O-GlcNAcylation in Caenorhabditis elegans. J Biol Chem. 2010; 285:2930–2939.
Article17. Lee J, Kim KY, Paik YK. Alteration in cellular acetylcholine influences dauer formation in Caenorhabditis elegans. BMB Rep. 2014; 47:80–85.
Article18. McGiven JA. New developments in the immunodiagnosis of brucellosis in livestock and wildlife. Rev Sci Tech. 2013; 32:163–176.
Article19. Pappas G. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents. 2010; 36:Suppl 1. S8–S11.20. Park WB, Im YB, Jung MH, Yoo HS. Molecular characteristics of Brucella abortus mutants generated using EZ-Tn5™ pMOD™-3 transposon system. J Prev Vet Med. 2015; 39:144–152.
Article21. Pizarro-Cerdá J, Méresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goñi I, Moreno E, Gorvel JP. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998; 66:5711–5724.
Article22. Pomastowski P, Buszewski B. Two-dimensional gel electrophoresis in the light of new developments. Trends Analyt Chem. 2014; 53:167–177.
Article23. Schlee S, Beinker P, Akhrymuk A, Reinstein J. A chaperone network for the resolubilization of protein aggregates: direct interaction of ClpB and DnaK. J Mol Biol. 2004; 336:275–285.
Article24. Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet Microbiol. 2010; 140:392–398.
Article25. Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004; 51:1525–1533.
Article26. Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011; 9:578–589.
Article27. Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004; 1694:67–80.
Article28. Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, Deutsch WA, Lenardo MJ. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007; 131:927–939.
Article29. Weldingh K, Rosenkrands I, Jacobsen S, Rasmussen PB, Elhay MJ, Andersen P. Two-dimensional electrophoresis for analysis of Mycobacterium tuberculosis culture filtrate and purification and characterization of six novel proteins. Infect Immun. 1998; 66:3492–3500.
Article30. Woods ML 2nd, Bonfiglioli R, McGee ZA, Georgopoulos C. Synthesis of a select group of proteins by Neisseria gonorrhoeae in response to thermal stress. Infect Immun. 1990; 58:719–725.
Article31. Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999; 274:28083–28086.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Sporozoite proteome analysis of Cryptosporidium parvum by one-dimensional SDS-PAGE and liquid chromatography tandem mass spectrometry
- Identification of Proteome Molecules by Proteomics Using Two-Dimensional Gel Electrophoresis and MALDI-TOF MS
- Different invasion efficiencies of Brucella abortus wild-type and mutantsin RAW 264.7 and THP-1 phagocytic cells and HeLa non-phagocytic cells
- 2DSpotDB: A Database for the Annotated Two-dimensional Polyacrylamide Gel Electrophoresis of Pathogen Proteins
- Protective effects of recombinant Brucella abortus Omp28 against infection with a virulent strain of Brucella abortus 544 in mice