J Vet Sci.
2013 Jun;14(2):107-114. 10.4142/jvs.2013.14.2.107.
Sporozoite proteome analysis of Cryptosporidium parvum by one-dimensional SDS-PAGE and liquid chromatography tandem mass spectrometry
- Affiliations
-
- 1Department of Veterinary Preclinical Science, Faculty of Veterinary Science, University of Liverpool, Liverpool L69 7ZJ, UK. zsiddiki@gmail.com
- KMID: 1705511
- DOI: http://doi.org/10.4142/jvs.2013.14.2.107
Abstract
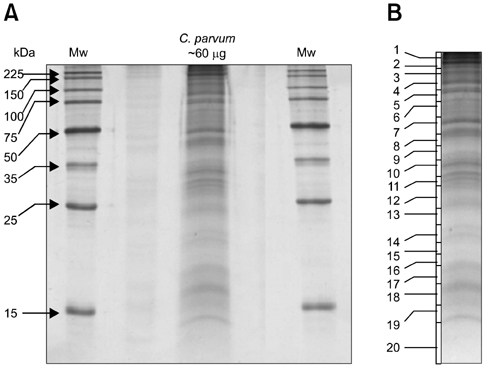
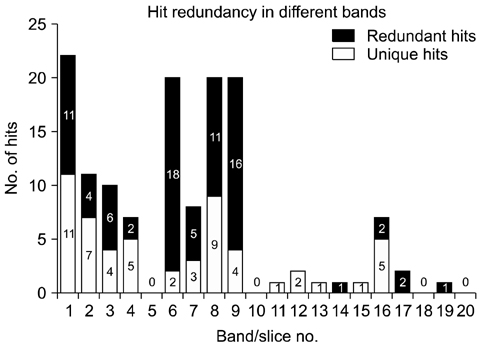
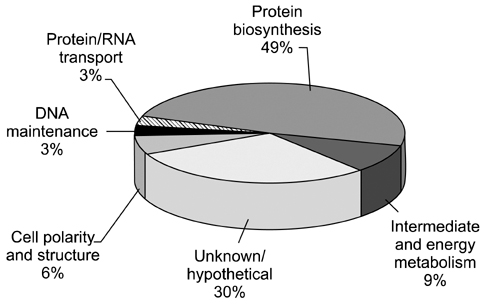
- Despite the development of new technologies, new challenges still remain for large scale proteomic profiling when dealing with complex biological mixtures. Fractionation prior to liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis is usually the preferred method to reduce the complexity of any biological sample. In this study, a gel LC-MS/MS approach was used to explore the stage specific proteome of Cryptosporidium (C.) parvum. To accomplish this, the sporozoite protein of C. parvum was first fractionated using SDS-PAGE with subsequent LC-MS/MS analysis. A total of 135 protein hits were recorded from 20 gel slices (from same gel lane), with many hits occurring in more than one band. Excluding all non-Cryptosporidium entries and proteins with multiple hits, 33 separate C. parvum entries were identified during the study. The overall goal of this study was to reduce sample complexity by protein fractionation and increase the possibility of detecting proteins present in lower abundance in a complex protein mixture.
MeSH Terms
-
Chemical Fractionation/methods
Chromatography, Liquid/methods/veterinary
Cryptosporidium parvum/*chemistry/growth & development/metabolism
Electrophoresis, Polyacrylamide Gel/methods/veterinary
Gene Expression Profiling/*methods/veterinary
Proteome/analysis
Proteomics/*methods
Protozoan Proteins/*analysis
Sporozoites/chemistry/metabolism
Tandem Mass Spectrometry/methods/veterinary
Proteome
Protozoan Proteins
Figure
Reference
-
1. Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004; 304:441–445.
Article2. Coombs GH. Biochemical peculiarities and drug targets in Cryptosporidium parvum: lessons from other coccidian parasites. Parasitol Today. 1999; 15:333–338.
Article3. Denton H, Brown SM, Roberts CW, Alexander J, McDonald V, Thong KW, Coombs GH. Comparison of the phosphofructokinase and pyruvate kinase activities of Cryptosporidium parvum, Eimeria tenella and Toxoplasma gondii. Mol Biochem Parasitol. 1996; 76:23–29.
Article4. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, Appella E, Hoffman SL, Yates JR 3rd, Carucci DJ, Sette A. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003; 100:9952–9957.
Article5. Entrala E, Mascaró C. Glycolytic enzyme activities in Cryptosporidium parvum oocysts. FEMS Microbiol Lett. 1997; 151:51–57.6. Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002; 419:520–526.
Article7. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002; 419:498–511.
Article8. Heiges M, Wang H, Robinson E, Aurrecoechea C, Gao X, Kaluskar N, Rhodes P, Wang S, He CZ, Su Y, Miller J, Kraemer E, Kissinger JC. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006; 34:D419–D422.9. Ivens A. Hurrah for genome projects! Parasitol Today. 2000; 16:317–320.
Article10. Lasonder E, Ishihama Y, Andersen JS, Vermunt AMW, Pain A, Sauerwein RW, Eling WMC, Hall N, Waters AP, Stunnenberg HG, Mann M. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002; 419:537–542.
Article11. O'Donoghue PJ. Cryptosporidium and cryptosporidiosis in man and animals. Int J Parasitol. 1995; 25:139–195.12. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999; 20:3551–3567.
Article13. Sam-Yellowe TY, Florens L, Wang T, Raine JD, Carucci DJ, Sinden R, Yates JR 3rd. Proteome analysis of rhoptry-enriched fractions isolated from Plasmodium merozoites. J Proteome Res. 2004; 3:995–1001.
Article14. Sanderson SJ, Xia D, Prieto H, Yates J, Heiges M, Kissinger JC, Bromley E, Lal K, Sinden RE, Tomley F, Wastling JM. Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics. 2008; 8:1398–1414.
Article15. Snelling WJ, Lin Q, Moore JE, Millar BC, Tosini F, Pozio E, Dooley JSG, Lowery CJ. Proteomics analysis and protein expression during sporozoite excystation of Cryptosporidium parvum (Coccidia, Apicomplexa). Mol Cell Proteomics. 2007; 6:346–355.
Article16. Thompson RCA, Olson ME, Zhu G, Enomoto S, Abrahamsen MS, Hijjawi NS. Cryptosporidium and cryptosporidiosis. Adv Parasitol. 2005; 59:77–158.17. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Geer LY, Helmberg W, Kapustin Y, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006; 34:D173–D180.
Article18. Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA. The genome of Cryptosporidium hominis. Nature. 2004; 431:1107–1112.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Charting the proteome of Cryptosporidium parvum sporozoites using sequence similarity-based BLAST searching
- Analysis of Low Molecular Weight Proteome from H. pylori Cell Extract Using the High Performance Liquid Chromatography
- Identification of Novel Metabolic Proteins Released by Insulin Signaling of the Rat Hypothalmus Using Liquid Chromatography-Mass Spectrometry (LC-MS)
- Metabolism and excretion of novel pulmonary-targeting docetaxel liposome in rabbits
- Application of Hemin-Agarose Affinity Chromatography to Enrich Proteome Components of Helicobacter pylori Strain 26695