J Vet Sci.
2018 Mar;19(2):188-199. 10.4142/jvs.2018.19.2.188.
In silico analysis of putative drug and vaccine targets of the metabolic pathways of Actinobacillus pleuropneumoniae using a subtractive/comparative genomics approach
- Affiliations
-
- 1Laboratory of Veterinary Pharmacokinetics and Pharmacodynamics, College of Veterinary Medicine, Kyungpook National University, Daegu 41566, Korea. parksch@knu.ac.kr
- 2Department of Chemistry Education, Teachers College, Kyungpook National University, Daegu 41566, Korea.
- KMID: 2407618
- DOI: http://doi.org/10.4142/jvs.2018.19.2.188
Abstract
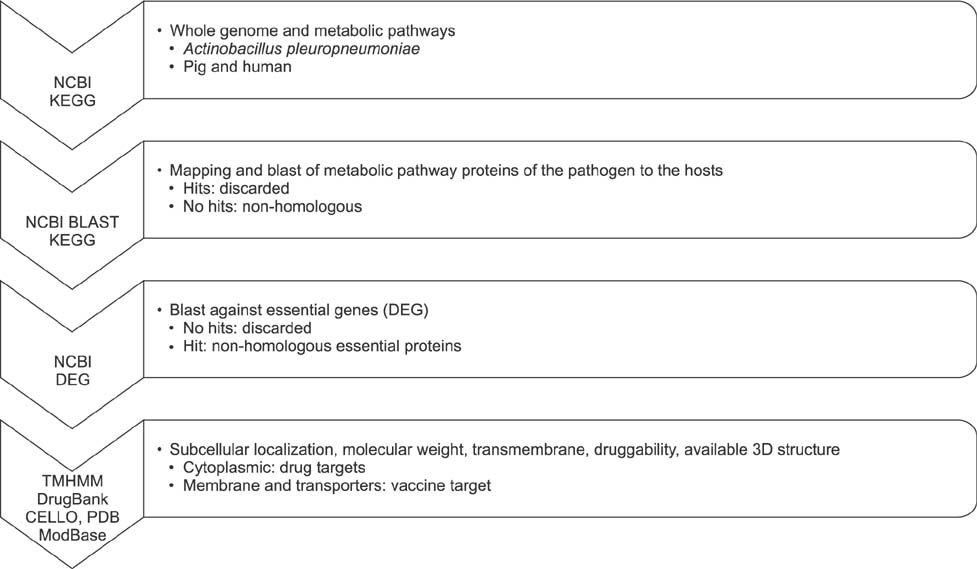
- Actinobacillus pleuropneumoniae is a Gram-negative bacterium that resides in the respiratory tract of pigs and causes porcine respiratory disease complex, which leads to significant losses in the pig industry worldwide. The incidence of drug resistance in this bacterium is increasing; thus, identifying new protein/gene targets for drug and vaccine development is critical. In this study, we used an in silico approach, utilizing several databases including the Kyoto Encyclopedia of Genes and Genomes (KEGG), the Database of Essential Genes (DEG), DrugBank, and Swiss-Prot to identify non-homologous essential genes and prioritize these proteins for their druggability. The results showed 20 metabolic pathways that were unique and contained 273 non-homologous proteins, of which 122 were essential. Of the 122 essential proteins, there were 95 cytoplasmic proteins and 11 transmembrane proteins, which are potentially suitable for drug and vaccine targets, respectively. Among these, 25 had at least one hit in DrugBank, and three had similarity to metabolic proteins from Mycoplasma hyopneumoniae, another pathogen causing porcine respiratory disease complex; thus, they could serve as common therapeutic targets. In conclusion, we identified glyoxylate and dicarboxylate pathways as potential targets for antimicrobial therapy and tetra-acyldisaccharide 4"²-kinase and 3-deoxy-D-manno-octulosonic-acid transferase as vaccine candidates against A. pleuropneumoniae.
Keyword
MeSH Terms
Figure
Reference
-
1. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000; 28:235–242.
Article2. Bossé JT, Janson H, Sheehan BJ, Beddek AJ, Rycroft AN, Kroll JS, Langford PR. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 2002; 4:225–235.
Article3. Butt AM, Nasrullah I, Tahir S, Tong Y. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS One. 2012; 7:e43080.4. Chawley P, Samal HB, Prava J, Suar M, Mahapatra RK. Comparative genomics study for identification of drug and vaccine targets in Vibrio cholerae: MurA ligase as a case study. Genomics. 2014; 103:83–93.
Article5. Chung WB, Bäckström LR, Collins MT. Experimental model of swine pneumonic pasteurellosis using crude Actinobacillus pleuropneumoniae cytotoxin and Pasteurella multocida given endobronchially. Can J Vet Res. 1994; 58:25–30.6. Damte D, Suh JW, Lee SJ, Yohannes SB, Hossain MA, Park SC. Putative drug and vaccine target protein identification using comparative genomic analysis of KEGG annotated metabolic pathways of Mycoplasma hyopneumoniae. Genomics. 2013; 102:47–56.
Article7. Doro F, Liberatori S, Rodríguez-Ortega MJ, Rinaudo CD, Rosini R, Mora M, Scarselli M, Altindis E, D'Aurizio R, Stella M, Margarit I, Maione D, Telford JL, Norais N, Grandi G. Surfome analysis as a fast track to vaccine discovery: identification of a novel protective antigen for Group B Streptococcus hypervirulent strain COH1. Mol Cell Proteomics. 2009; 8:1728–1737.8. Doytchinova IA, Flower DR. Bioinformatic approach for identifying parasite and fungal candidate subunit vaccines. Open Vaccine J. 2008; 1:22–26.
Article9. Duffield M, Cooper I, McAlister E, Bayliss M, Ford D, Oyston P. Predicting conserved essential genes in bacteria: in silico identification of putative drug targets. Mol Biosyst. 2010; 6:2482–2489.
Article10. Garrett TA, Que NL, Raetz CR. Accumulation of a lipid A precursor lacking the 4′-phosphate following inactivation of the Escherichia coli lpxK gene. J Biol Chem. 1998; 273:12457–12465.
Article11. Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol Cell. 2013; 52:248–254.
Article12. Glory E, Murphy RF. Automated subcellular location determination and high-throughput microscopy. Dev Cell. 2007; 12:7–16.
Article13. Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998; 62:1079–1093.
Article14. Gottschalk M. Actinobacillosis. In : Zimmerman JJ, Karriker LA, Schwartz KJ, Stevenson GW, editors. Diseases of Swine. 10th ed. Chichester: Wiley-Blackwell;2012. p. 653–669.15. Huang H, Potter AA, Campos M, Leighton FA, Willson PJ, Haines DM, Yates WD. Pathogenesis of porcine Actinobacillus pleuropneumonia, part II: roles of proinflammatory cytokines. Can J Vet Res. 1999; 63:69–78.16. Hung MC, Link W. Protein localization in disease and therapy. J Cell Sci. 2011; 124:3381–3392.
Article17. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016; 44:D457–D462.
Article18. Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun. 2013; 4:3001.
Article19. Kim B, Min K, Choi C, Cho WS, Cheon DS, Kwon D, Kim J, Chae C. Antimicrobial susceptibility of Actinobacillus pleuropneumoniae isolated from pigs in Korea using new standardized procedures. J Vet Med Sci. 2001; 63:341–342.
Article20. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001; 305:567–580.
Article21. Li C, Ye Z, Wen L, Chen R, Tian L, Zhao F, Pan J. Identification of a novel vaccine candidate by immunogenic screening of Vibrio parahaemolyticus outer membrane proteins. Vaccine. 2014; 32:6115–6121.
Article22. Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014; 42:D574–D580.23. Mobegi FM, van Hijum SA, Burghout P, Bootsma HJ, de Vries SP, van der Gaast-de Jongh CE, Simonetti E, Langereis JD, Hermans PW, de Jonge MI, Zomer A. From microbial gene essentiality to novel antimicrobial drug targets. BMC Genomics. 2014; 15:958.
Article24. Morya VK, Dewaker V, Mecarty SD, Singh R. In silico analysis metabolic pathways for identification of putative drug targets for Staphylococcus aureus. J Comput Sci Syst Biol. 2010; 3:62–69.25. Nguyen-Distèche M, Fraipont C, Buddelmeijer N, Nanninga N. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol Life Sci. 1998; 54:309–316.
Article26. Nishio M, Okada N, Miki T, Haneda T, Danbara H. Identification of the outer-membrane protein PagC required for the serum resistance phenotype in Salmonella enterica serovar Choleraesuis. Microbiology. 2005; 151:863–873.
Article27. Opriessnig T, Giménez-Lirola LG, Halbur PG. Polymicrobial respiratory disease in pigs. Anim Health Res Rev. 2011; 12:133–148.
Article28. Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA, Nilges M, Sali A. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2014; 42:D336–D346.
Article29. Pinho MG, Filipe SR, de Lencastre H, Tomasz A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol. 2001; 183:6525–6531.
Article30. Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008; 62:35–51.
Article31. Seib KL, Dougan G, Rappuoli R. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet. 2009; 5:e1000612.
Article32. Shanmugham B, Pan A. Identification and characterization of potential therapeutic candidates in emerging human pathogen Mycobacterium abscessus: a novel hierarchical in silico approach. PLoS One. 2013; 8:e59126.33. Simon I, Wright M, Flohr T, Hevezi P, Caras IW. Determining subcellular localization of novel drug targets by transient transfection in COS cells. Cytotechnology. 2001; 35:189–196.34. Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975; 72:2999–3003.
Article35. Uddin R, Saeed K, Khan W, Azam SS, Wadood A. Metabolic pathway analysis approach: identification of novel therapeutic target against methicillin resistant Staphylococcus aureus. Gene. 2015; 556:213–226.
Article36. UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212.37. Vanni M, Merenda M, Barigazzi G, Garbarino C, Luppi A, Tognetti R, Intorre L. Antimicrobial resistance of Actinobacillus pleuropneumoniae isolated from swine. Vet Microbiol. 2012; 156:172–177.
Article38. Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006; 34:D668–D672.39. Wu XB, Tian LH, Zou HJ, Wang CY, Yu ZQ, Tang CH, Zhao FK, Pan JY. Outer membrane protein OmpW of Escherichia coli is required for resistance to phagocytosis. Res Microbiol. 2013; 164:848–855.
Article40. Yoo AN, Cha SB, Shin MK, Won HK, Kim EH, Choi HW, Yoo HS. Serotypes and antimicrobial resistance patterns of the recent Korean Actinobacillus pleuropneumoniae isolates. Vet Rec. 2014; 174:223.
Article41. Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006; 64:643–651.
Article42. Zhang R, Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009; 37:D455–D458.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevalence and Characterization of Actinobacillus pleuropneumoniae Isolated from Korean Pigs
- Antibiotic resistance in Neisseria gonorrhoeae: broad-spectrum drug target identification using subtractive genomics
- Generation of transgenic corn-derived Actinobacillus pleuropneumoniae ApxIIA fused with the cholera toxin B subunit as a vaccine candidate
- Development of Actinobacillus pleuropneumoniae ApxI, ApxII, and ApxIII-specific ELISA methods for evaluation of vaccine efficiency
- Multi-epitope vaccine against drug-resistant strains of Mycobacterium tuberculosis: a proteome-wide subtraction and immunoinformatics approach