Ann Lab Med.
2018 Mar;38(2):160-164. 10.3343/alm.2018.38.2.160.
Clinicopathological Characteristics of Hyperdiploidy with High-Risk Cytogenetics in Multiple Myeloma
- Affiliations
-
- 1Department of Laboratory Medicine, College of Medicine, Ewha Womans University, Seoul, Korea. JungWonH@ewha.ac.kr
- 2Department of Internal Medicine, College of Medicine, Ewha Womans University, Seoul, Korea.
- 3Department of Laboratory Medicine, Dongguk University, Ilsan Medical Center, Goyang, Korea. hjhuh@duih.org
- KMID: 2403362
- DOI: http://doi.org/10.3343/alm.2018.38.2.160
Abstract
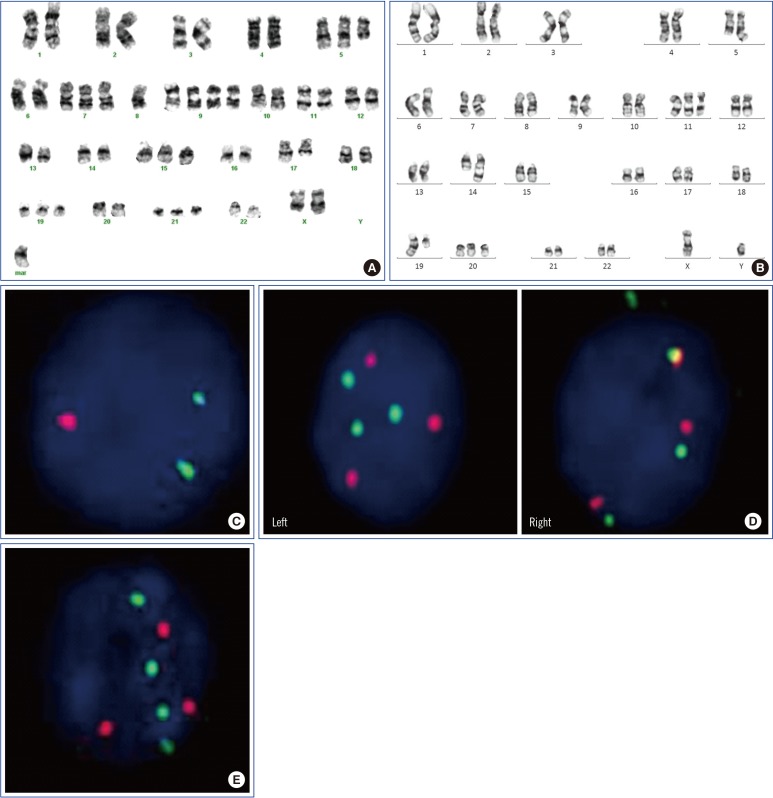
- In multiple myeloma (MM), hyperdiploidy (HD) is known to impart longer overall survival. However, it is unclear whether coexistent HD ameliorates the adverse effects of known high-risk cytogenetics in MM patients. To address this issue, we investigated the clinicopathological characteristics of HD with high-risk cytogenetics in MM. Ninety-seven patients with MM were included in the study. For metaphase cytogenetics (MC), unstimulated cells from bone marrow aspirates were cultured for either 24 or 48 hours. To detect HD by interphase fluorescence in situ hybridization (iFISH), we assessed trisomies of chromosomes 5, 7, 9, 11, 15, and 17. Of the 97 MM patients, 40 showed HD. The frequency of co-occurrence of HD and high-risk cytogenetics was 14% (14/97). When the clinicopathological characteristics were compared between the two groups of HD with high-risk cytogenetics vs. non-HD (NHD) with high-risk cytogenetics, the level of beta 2 microglobulin and stage distribution significantly differed (P=0.020, P=0.032, respectively). This study shows that some of the clinicopathological characteristics of MM patients with high-risk cytogenetics differ according to HD or NHD status.
Keyword
MeSH Terms
Figure
Reference
-
1. Rajkumar SV. Myeloma today: disease definitions and treatment advances. Am J Hematol. 2016; 91:90–100. PMID: 26565896.2. Stella F, Pedrazzini E, Agazzoni M, Ballester O, Slavutsky I. Cytogenetic alterations in multiple myeloma: prognostic significance and the choice of frontline therapy. Cancer Invest. 2015; 33:496–504. PMID: 26506456.3. Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23:2210–2221. PMID: 19798094.4. Carballo-Zarate AA, Medeiros LJ, Fang L, Shah JJ, Weber DM, Thomas SK, et al. Additional-structural-chromosomal aberrations are associated with inferior clinical outcome in patients with hyperdiploid multiple myeloma: a single-institution experience. Mod Pathol. 2017; 30:843–853. PMID: 28281554.5. Hebraud B, Magrangeas F, Cleynen A, Lauwers-Cances V, Chretien ML, Hulin C, et al. Role of additional chromosomal changes in the prognostic value of t(4;14) and del(17p) in multiple myeloma: the IFM experience. Blood. 2015; 125:2095–2100. PMID: 25636340.6. Kumar S, Fonseca R, Ketterling RP, Dispenzieri A, Lacy MQ, Gertz MA, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012; 119:2100–2105. PMID: 22234687.7. Merz M, Hielscher T, Seckinger A, Hose D, Mai EK, Raab MS, et al. Baseline characteristics, chromosomal alterations, and treatment affecting prognosis of deletion 17p in newly diagnosed myeloma. Am J Hematol. 2016; 91:E473–E477. PMID: 27508939.8. Pawlyn C, Melchor L, Murison A, Wardell CP, Brioli A, Boyle EM, et al. Coexistent hyperdiploidy does not abrogate poor prognosis in myeloma with adverse cytogenetics and may precede IGH translocations. Blood. 2015; 125:831–840. PMID: 25428216.9. International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003; 121:749–757. PMID: 12780789.10. Shaffer LG, McGowan-Jordan J, Schmid M. ISCN 2013: an international system for human cytogenetic nomenclature (2013): Karger Medical and Scientific Publishers. 2013.11. Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised International Staging System for multiple myeloma: a report from International Myeloma Working Group. J Clin Oncol. 2015; 33:2863–2869. PMID: 26240224.12. Chretien ML, Corre J, Lauwers-Cances V, Magrangeas F, Cleynen A, Yon E, et al. Understanding the role of hyperdiploidy in myeloma prognosis: which trisomies really matter? Blood. 2015; 126:2713–2719. PMID: 26516228.13. Lim JH, Seo EJ, Park CJ, Jang S, Chi HS, Suh C, et al. Cytogenetic classification in Korean multiple myeloma patients: prognostic significance of hyperdiploidy with 47-50 chromosomes and the number of structural abnormalities. Eur J Haematol. 2014; 92:313–320. PMID: 24372944.14. Ortega M, Mallo M, Sole F, Sanchez-Morata C, Lopez-Andreoni L, Martinez-Morgado N, et al. 5q- syndrome and multiple myeloma diagnosed simultaneously and successful treated with lenalidomide. Leuk Res. 2013; 37:1248–1250. PMID: 23891188.15. Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007; 110:719–726. PMID: 17420284.16. Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015; 33:3911–3920. PMID: 26282654.17. Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci U S A. 2011; 108:632–637. PMID: 21187408.18. Wirth M, Stojanovic N, Christian J, Paul MC, Stauber RH, Schmid RM, et al. MYC and EGR1 synergize to trigger tumor cell death by controlling NOXA and BIM transcription upon treatment with the proteasome inhibitor bortezomib. Nucleic Acids Res. 2014; 42:10433–10447. PMID: 25147211.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A retrospective analysis of cytogenetic alterations in patients with newly diagnosed multiple myeloma: a single center study in Korea
- Multiple myeloma
- Amylase-Producing IgG Kappa Type Multiple Myeloma
- Differing Outcomes of Patients with High Hyperdiploidy and ETV6-RUNX1 Rearrangement in Korean Pediatric Precursor B Cell Acute Lymphoblastic Leukemia
- Rapid Progression of Solitary Plasmacytoma to Multiple Myeloma in Lumbar Vertebra