Blood Res.
2016 Jun;51(2):122-126. 10.5045/br.2016.51.2.122.
A retrospective analysis of cytogenetic alterations in patients with newly diagnosed multiple myeloma: a single center study in Korea
- Affiliations
-
- 1Department of Laboratory Medicine, Dong-A University College of Medicine, Busan, Korea. jyhan@dau.ac.kr
- 2Department of Internal Medicine, Dong-A University College of Medicine, Busan, Korea.
- KMID: 2309214
- DOI: http://doi.org/10.5045/br.2016.51.2.122
Abstract
- BACKGROUND
The accurate identification of cytogenetic abnormalities in multiple myeloma (MM) has become more important over recent years for the development of new diagnostic and prognostic markers. In this study, we retrospectively analyzed the cytogenetic aberrations in MM cases as an initial assessment in a single institute.
METHODS
We reviewed the cytogenetic results from 222 patients who were newly diagnosed with MM between January 2000 and December 2015. Chromosomal analysis was performed on cultured bone marrow samples by standard G-banding technique. At least 20 metaphase cells were analyzed for karyotyping.
RESULTS
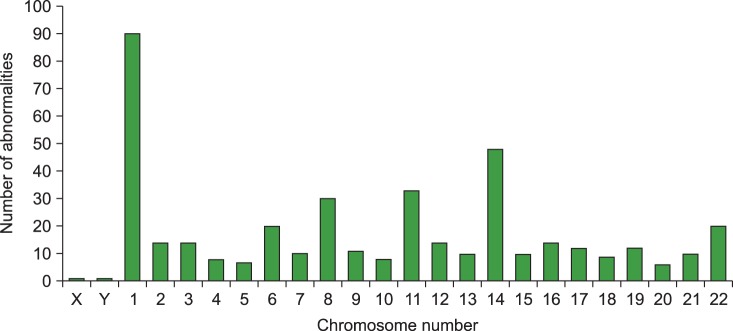
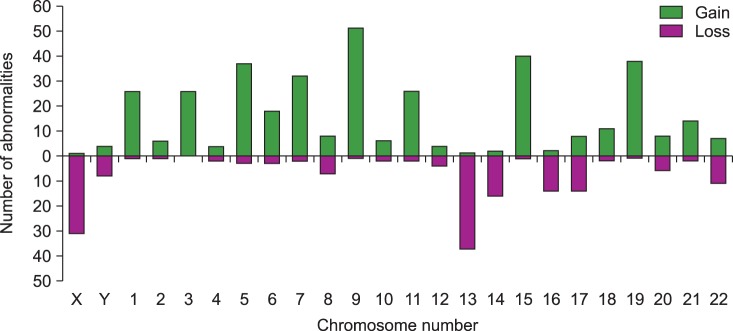
Clonal chromosome abnormalities were detected in 45.0% (100/222) of the patients. Among these results, 80 cases (80.0%) had both numerical and structural chromosome abnormalities. Overall hyperdiploidy with structural cytogenetic aberrations was the most common finding (44.0%), followed by hypodiploidy with structural aberrations (28.0%). Amplification of the long arm of chromosome 1 and -13/del(13q) were the most frequent recurrent abnormalities, and were detected in 50 patients (50.0%) and 40 patients (40.0%) with clonal abnormalities, respectively. The most common abnormality involving 14q32 was t(11;14)(q13;q32), which was observed in 19 cases.
CONCLUSION
These findings demonstrate that myeloma cells exhibit complex aberrations regardless of ploidy, even from a single center in Korea. Conventional cytogenetic analysis should be included in the initial diagnostic work-up for patients suspected of having MM.
MeSH Terms
Figure
Reference
-
1. Rajkumar SV. Multiple myeloma: 2014 Update on diagnosis, risk-stratification, and management. Am J Hematol. 2014; 89:999–1009. PMID: 25223428.
Article2. Talley PJ, Chantry AD, Buckle CH. Genetics in myeloma: genetic technologies and their application to screening approaches in myeloma. Br Med Bull. 2015; 113:15–30. PMID: 25662536.
Article3. Munshi NC, Anderson KC, Bergsagel PL, et al. Consensus recommendations for risk stratification in multiple myeloma: report of the International Myeloma Workshop Consensus Panel 2. Blood. 2011; 117:4696–4700. PMID: 21292777.
Article4. Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014; 15:e538–e548. PMID: 25439696.5. Viguié F. Multiple myeloma. Atlas Genet Cytogenet Oncol Haematol. 2004; 8:255–257.
Article6. Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011; 204:3–12. PMID: 21356186.
Article7. Jimenez-Zepeda VH, Braggio E, Fonseca R. Dissecting karyotypic patterns in non-hyperdiploid multiple myeloma: an overview on the karyotypic evolution. Clin Lymphoma Myeloma Leuk. 2013; 13:552–558. PMID: 23856591.
Article8. Shaffer LG, McGowan-Jordan J, Schmid M. An international system for human cytogenetic nomenclature. Basel, Switzerland: S Karger;2013.9. Greipp PR, San Miguel J, Durie BG, et al. International staging system for multiple myeloma. J Clin Oncol. 2005; 23:3412–3420. PMID: 15809451.
Article10. Lai YY, Huang XJ, Cai Z, et al. Prognostic power of abnormal cytogenetics for multiple myeloma: a multicenter study in China. Chin Med J (Engl). 2012; 125:2663–2670. PMID: 22931972.11. Lim AS, Lim TH, See KH, et al. Cytogenetic and molecular aberrations of multiple myeloma patients: a single-center study in Singapore. Chin Med J (Engl). 2013; 126:1872–1877. PMID: 23673102.12. Kumar S, Fonseca R, Ketterling RP, et al. Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood. 2012; 119:2100–2105. PMID: 22234687.
Article13. Berry NK, Bain NL, Enjeti AK, Rowlings P. Genomic profiling of plasma cell disorders in a clinical setting: integration of microarray and FISH, after CD138 selection of bone marrow. J Clin Pathol. 2014; 67:66–69. PMID: 23969274.
Article14. Waheed S, Shaughnessy JD, van Rhee F, et al. International staging system and metaphase cytogenetic abnormalities in the era of gene expression profiling data in multiple myeloma treated with total therapy 2 and 3 protocols. Cancer. 2011; 117:1001–1009. PMID: 20945320.
Article15. Jekarl DW, Min CK, Kwon A, et al. Impact of genetic abnormalities on the prognoses and clinical parameters of patients with multiple myeloma. Ann Lab Med. 2013; 33:248–254. PMID: 23826560.
Article16. Lim JH, Seo EJ, Park CJ, et al. Cytogenetic classification in Korean multiple myeloma patients: prognostic significance of hyperdiploidy with 47-50 chromosomes and the number of structural abnormalities. Eur J Haematol. 2014; 92:313–320. PMID: 24372944.
Article17. Kim SH, Kim JH, Lee DM, et al. Comparison between conventional cytogenetics and interphase fluorescence in situ hybridization (FISH) for patients with multiple myeloma. Korean J Hematol. 2009; 44:14–21.
Article18. Jimenez-Zepeda VH, Neme-Yunes Y, Braggio E. Chromosome abnormalities defined by conventional cytogenetics in plasma cell leukemia: what have we learned about its biology? Eur J Haematol. 2011; 87:20–27. PMID: 21692850.
Article19. Oh S, Koo DH, Kwon MJ, et al. Chromosome 13 deletion and hypodiploidy on conventional cytogenetics are robust prognostic factors in Korean multiple myeloma patients: web-based multicenter registry study. Ann Hematol. 2014; 93:1353–1361. PMID: 24671365.
Article20. Caltagirone S, Ruggeri M, Aschero S, et al. Chromosome 1 abnormalities in elderly patients with newly diagnosed multiple myeloma treated with novel therapies. Haematologica. 2014; 99:1611–1617. PMID: 25015938.
Article21. An G, Xu Y, Shi L, et al. Chromosome 1q21 gains confer inferior outcomes in multiple myeloma treated with bortezomib but copy number variation and percentage of plasma cells involved have no additional prognostic value. Haematologica. 2014; 99:353–359. PMID: 24213147.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Correlation of Chromosomal Aberrations with Prognostic Markers in Multiple Myeloma Patients- A Single Institution Study
- Acute Pancreatitis with Hypercalcemia as Initial Manifestation of Multiple Myeloma
- Clinical Utility of a Diagnostic Approach to Detect Genetic Abnormalities in Multiple Myeloma: A Single Institution Experience
- Newly Developed Multiple Myeloma in a Patient with Primary T-Cell Lymphoma of Bone
- Clinical significance of chromosomal abnormality in multiple myeloma