Immune Netw.
2017 Apr;17(2):77-88. 10.4110/in.2017.17.2.77.
Mitochondrial Control of Innate Immunity and Inflammation
- Affiliations
-
- 1Department of Microbiology, Chungnam National University School of Medicine, Daejeon 35015, Korea. hayoungj@cnu.ac.kr
- 2Infection Signaling Network Research Center, Chungnam National University School of Medicine, Daejeon 35015, Korea.
- 3Biomedical Research Institute, Chungnam National University Hospital, Daejeon 35015, Korea.
- 4Center for Convergent Research of Emerging Virus Infection, Korea Research Institute of Chemical Technology, Daejeon 34114, Korea.
- KMID: 2376865
- DOI: http://doi.org/10.4110/in.2017.17.2.77
Abstract
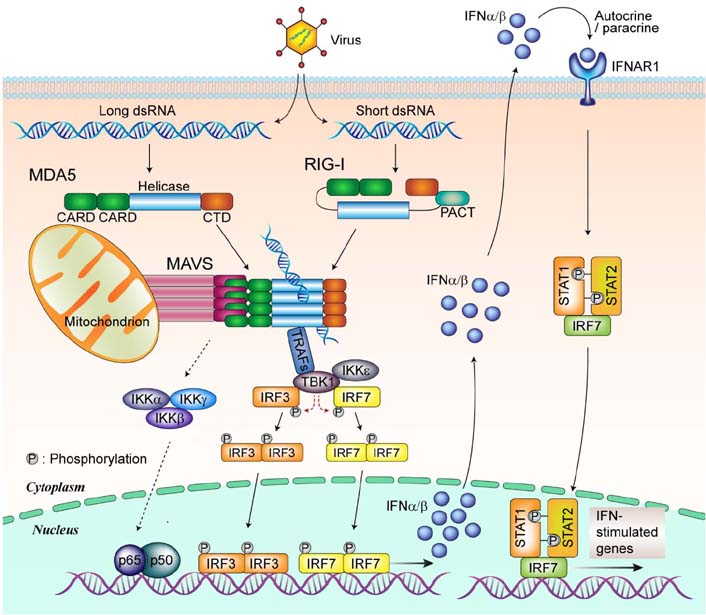
- Mitochondria are key organelles involved in energy production, functioning as the metabolic hubs of cells. Recent findings emphasize the emerging role of the mitochondrion as a key intracellular signaling platform regulating innate immune and inflammatory responses. Several mitochondrial proteins and mitochondrial reactive oxygen species have emerged as central players orchestrating the innate immune responses to pathogens and damaging ligands. This review explores our current understanding of the roles played by mitochondria in regulation of innate immunity and inflammatory responses. Recent advances in our understanding of the relationship between autophagy, mitochondria, and inflammasome activation are also briefly discussed. A comprehensive understanding of mitochondrial role in toll-like receptor-mediated innate immune responses and NLRP3 inflammasome complex activation, will facilitate development of novel therapeutics to treat various infectious, inflammatory, and autoimmune disorders.
Keyword
MeSH Terms
Figure
Cited by 3 articles
-
The autophagy Protein Atg5 Plays a Crucial Role in the Maintenance and Reconstitution Ability of Hematopoietic Stem Cells
Hi Eun Jung, Ye Ri Shim, Ji Eun Oh, Dong Sun Oh, Heung Kyu Lee
Immune Netw. 2019;19(2):. doi: 10.4110/in.2019.19.e12.Damage-Associated Molecular Patterns in Inflammatory Diseases
Jong Seong Roh, Dong Hyun Sohn
Immune Netw. 2018;18(4):e27. doi: 10.4110/in.2018.18.e27.Damage-Associated Molecular Patterns in Inflammatory Diseases
Jong Seong Roh, Dong Hyun Sohn
Immune Netw. 2018;18(4):. doi: 10.4110/in.2018.18.e27.
Reference
-
1. Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal. 2013; 19:240–242.
Article2. Taylor DE, Ghio AJ, Piantadosi CA. Reactive oxygen species produced by liver mitochondria of rats in sepsis. Arch Biochem Biophys. 1995; 316:70–76.
Article3. Kurose I, Miura S, Fukumura D, Yonei Y, Saito H, Tada S, Suematsu M, Tsuchiya M. Nitric oxide mediates Kupffer cell-induced reduction of mitochondrial energization in hepatoma cells: a comparison with oxidative burst. Cancer Res. 1993; 53:2676–2682.4. Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem. 1992; 267:5317–5323.
Article5. Cloonan SM, Choi AM. Mitochondria: sensors and mediators of innate immune receptor signaling. Curr Opin Microbiol. 2013; 16:327–338.
Article6. Dromparis P, Michelakis ED. Mitochondria in vascular health and disease. Annu Rev Physiol. 2013; 75:95–126.
Article7. Gurung P, Lukens JR, Kanneganti TD. Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med. 2015; 21:193–201.
Article8. Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015; 42:406–417.
Article9. Lopez-Armada MJ, Riveiro-Naveira RR, Vaamonde-Garcia C, Valcarcel-Ares MN. Mitochondrial dysfunction and the inflammatory response. Mitochondrion. 2013; 13:106–118.
Article10. Sandhir R, Halder A, Sunkaria A. Mitochondria as a centrally positioned hub in the innate immune response. Biochim Biophys Acta. 2016; DOI: 10.1016/j.bbadis.2016.10.020.
Article11. Kim MJ, Yoon JH, Ryu JH. Mitophagy: a balance regulator of NLRP3 inflammasome activation. BMB Rep. 2016; 49:529–535.
Article12. Kugelberg E. Immunometabolism: Mitochondria adapt to bacteria. Nat Rev Immunol. 2016; 16:464–465.13. Wasilewski M, Chojnacka K, Chacinska A. Protein trafficking at the crossroads to mitochondria. Biochim Biophys Acta. 2017; 1864:125–137.
Article14. Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015; 16:530–542.
Article15. Hekimi S, Wang Y, Noe A. Mitochondrial ROS and the effectors of the intrinsic apoptotic pathway in aging cells: The discerning killers! Front Genet. 2016; 7:161.
Article16. Dan Dunn J, Alvarez LA, Zhang X, Soldati T. Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox Biol. 2015; 6:472–485.
Article17. Rimessi A, Previati M, Nigro F, Wieckowski MR, Pinton P. Mitochondrial reactive oxygen species and inflammation: Molecular mechanisms, diseases and promising therapies. Int J Biochem Cell Biol. 2016; 81:281–293.
Article18. Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012; 337:1062–1065.
Article19. Haroon S, Vermulst M. Linking mitochondrial dynamics to mitochondrial protein quality control. Curr Opin Genet Dev. 2016; 38:68–74.
Article20. Tal MC, Iwasaki A. Mitoxosome: a mitochondrial platform for cross-talk between cellular stress and antiviral signaling. Immunol Rev. 2011; 243:215–234.
Article21. Brubaker SW, Iwasaki A. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015; 33:257–290.
Article22. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010; 11:373–384.
Article23. Skevaki C, Pararas M, Kostelidou K, Tsakris A, Routsias JG. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious diseases. Clin Exp Immunol. 2015; 180:165–177.
Article24. Yarovinsky F, Zhang D, Andersen JF, Bannenberg GL, Serhan CN, Hayden MS, Hieny S, Sutterwala FS, Flavell RA, Ghosh S, Sher A. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 2005; 308:1626–1629.
Article25. Zhang D, Zhang G, Hayden MS, Greenblatt MB, Bussey C, Flavell RA, Ghosh S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science. 2004; 303:1522–1526.
Article26. Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006; 442:39–44.
Article27. Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011; 34:665–679.
Article28. Yuk JM, Jo EK. Toll-like Receptors and Innate Immunity. J Bacteriol Virol. 2011; 41:225–235.
Article29. Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012; 481:278–286.
Article30. Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012; 13:325–332.
Article31. Tall AR, Yvan-Charvet L. Cholesterol, inflammation and innate immunity. Nat Rev Immunol. 2015; 15:104–116.
Article32. Gombault A, Baron L, Couillin I. ATP release and purinergic signaling in NLRP3 inflammasome activation. Front Immunol. 2012; 3:414.
Article33. Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014; 156:1193–1206.
Article34. Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: accomplices and antagonists of antiviral signal transduction. J Virol. 2014; 88:8194–8200.
Article35. Chan YK, Gack MU. RIG-I-like receptor regulation in virus infection and immunity. Curr Opin Virol. 2015; 12:7–14.
Article36. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007; 446:916–920.
Article37. Lang X, Tang T, Jin T, Ding C, Zhou R, Jiang W. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med. 2017; 214:459–473.
Article38. Eisenacher K, Krug A. Regulation of RLR-mediated innate immune signaling--it is all about keeping the balance. Eur J Cell Biol. 2012; 91:36–47.39. Thaiss CA, Levy M, Itav S, Elinav E. Integration of Innate Immune Signaling. Trends Immunol. 2016; 37:84–101.
Article40. Li M, Zhou Y, Feng G, Su SB. The critical role of Toll-like receptor signaling pathways in the induction and progression of autoimmune diseases. Curr Mol Med. 2009; 9:365–374.
Article41. Vazquez C, Horner SM. MAVS coordination of antiviral innate immunity. J Virol. 2015; 89:6974–6977.
Article42. Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011; 23:564–572.
Article43. Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005; 122:669–682.
Article44. Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006; 24:633–642.
Article45. Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011; 146:448–461.
Article46. Cheng G, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci U S A. 2007; 104:9035–9040.
Article47. Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2008; 105:14046–14051.
Article48. Buss C, Opitz B, Hocke AC, Lippmann J, van Laak V, Hippenstiel S, Krüll M, Suttorp N, Eitel J. Essential role of mitochondrial antiviral signaling, IFN regulatory factor (IRF)3, and IRF7 in Chlamydophila pneumoniae-mediated IFN-beta response and control of bacterial replication in human endothelial cells. J Immunol. 2010; 184:3072–3078.49. Li XD, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci U S A. 2011; 108:17390–17395.
Article50. Castanier C, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010; 11:133–138.
Article51. Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013; 153:348–361.
Article52. El Maadidi S, Faletti L, Berg B, Wenzl C, Wieland K, Chen ZJ, Maurer U, Borner C. A novel mitochondrial MAVS/Caspase-8 platform links RNA virus-induced innate antiviral signaling to Bax/Bak-independent apoptosis. J Immunol. 2014; 192:1171–1183.
Article53. Huang Y, Liu H, Li S, Tang Y, Wei B, Yu H, Wang C. MAVS-MKK7-JNK2 defines a novel apoptotic signaling pathway during viral infection. PLoS Pathog. 2014; 10:e1004020.
Article54. Liehl P, Zuzarte-Luis V, Chan J, Zillinger T, Baptista F, Carapau D, Konert M, Hanson KK, Carret C, Lassnig C, Müller M, Kalinke U, Saeed M, Chora AF, Golenbock DT, Strobl B, Prudencio M, Coelho LP, Kappe SH, Superti-Furga G, Pichlmair A, Vigário AM, Rice CM, Fitzgerald KA, Barchet W, Mota MM. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat Med. 2014; 20:47–53.
Article55. Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006; 25:3257–3263.
Article56. Zhou Z, Jia X, Xue Q, Dou Z, Ma Y, Zhao Z, Jiang Z, He B, Jin Q, Wang J. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc Natl Acad Sci U S A. 2014; 111:E245–E254.
Article57. You F, Wang P, Yang L, Yang G, Zhao YO, Qian F, Walker W, Sutton R, Montgomery R, Lin R, Iwasaki A, Fikrig E. ELF4 is critical for induction of type I interferon and the host antiviral response. Nat Immunol. 2013; 14:1237–1246.
Article58. Song T, Wei C, Zheng Z, Xu Y, Cheng X, Yuan Y, Guan K, Zhang Y, Ma Q, Shi W, Zhong H. c-Abl tyrosine kinase interacts with MAVS and regulates innate immune response. FEBS Lett. 2010; 584:33–38.
Article59. Liu S, Wei C, Zheng Z, Xu Y, Cheng X, Yuan Y, Guan K, Zhang Y, Ma Q, Shi W, Zhong H. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013; 2:e00785.
Article60. Cao Z, Zhou Y, Zhu S, Feng J, Chen X, Liu S, Peng N, Yang X, Xu G, Zhu Y. Pyruvate Carboxylase Activates the RIG-I-like Receptor-Mediated Antiviral Immune Response by Targeting the MAVS signalosome. Sci Rep. 2016; 6:22002.
Article61. You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009; 10:1300–1308.
Article62. Zhou X, You F, Chen H, Jiang Z. Poly(C)-binding protein 1 (PCBP1) mediates housekeeping degradation of mitochondrial antiviral signaling (MAVS). Cell Res. 2012; 22:717–727.
Article63. Jia Y, Song T, Wei C, Ni C, Zheng Z, Xu Q, Ma H, Li L, Zhang Y, He X, Xu Y, Shi W, Zhong H. Negative regulation of MAVS-mediated innate immune response by PSMA7. J Immunol. 2009; 183:4241–4248.
Article64. Allen IC, Song T, Wei C, Ni C, Zheng Z, Xu Q, Ma H, Li L, Zhang Y, He X, Xu Y, Shi W, Zhong H. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011; 34:854–865.
Article65. Soares F, Tattoli I, Wortzman ME, Arnoult D, Philpott DJ, Girardin SE. NLRX1 does not inhibit MAVS-dependent antiviral signalling. Innate Immun. 2013; 19:438–448.
Article66. Zhao Y, Sun X, Nie X, Sun L, Tang TS, Chen D, Sun Q. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLoS Pathog. 2012; 8:e1003086.
Article67. Wang P, Yang L, Cheng G, Yang G, Xu Z, You F, Sun Q, Lin R, Fikrig E, Sutton RE. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 2013; 3:1057–1070.
Article68. Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol. 2014; 192:4758–4764.
Article69. Shi Y, Yuan B, Qi N, Zhu W, Su J, Li X, Qi P, Zhang D, Hou F. An autoinhibitory mechanism modulates MAVS activity in antiviral innate immune response. Nat Commun. 2015; 6:7811.
Article70. Chan YK, Gack MU. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol. 2016; 17:523–530.
Article71. Xia P, Wang S, Xiong Z, Ye B, Huang LY, Han ZG, Fan Z. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun. 2015; 6:8132.
Article72. Yoo YS, Park YY, Kim JH, Cho H, Kim SH, Lee HS, Kim TH, Kim YS, Lee Y, Kim CJ, Jung JU, Lee JS, Cho H. The mitochondrial ubiquitin ligase MARCH5 resolves MAVS aggregates during antiviral signalling. Nat Commun. 2015; 6:7910.
Article73. Xiang W, Zhang Q, Lin X, Wu S, Zhou Y, Meng F, Fan Y, Shen T, Xiao M, Xia Z, Zou J, Feng XH, Xu P. PPM1A silences cytosolic RNA sensing and antiviral defense through direct dephosphorylation of MAVS and TBK1. Sci Adv. 2016; 2:e1501889.
Article74. Choi YB, Shembade N, Parvatiyar K, Balachandran S, Harhaj EW. TAX1BP1 Restrains Virus-Induced Apoptosis by Facilitating Itch-Mediated Degradation of the Mitochondrial Adaptor MAVS. Mol Cell Biol. 2017; 37:pii: e00422-16.
Article75. Shi HX, Liu X, Wang Q, Tang PP, Liu XY, Shan YF, Wang C. Mitochondrial ubiquitin ligase MARCH5 promotes TLR7 signaling by attenuating TANK action. PLoS Pathog. 2011; 7:e1002057.
Article76. West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011; 472:476–480.
Article77. Geng J, Sun X, Wang P, Zhang S, Wang X, Wu H, Hong L, Xie C, Li X, Zhao H, Liu Q, Jiang M, Chen Q, Zhang J, Li Y, Song S, Wang HR, Zhou R, Johnson RL, Chien KY, Lin SC, Han J, Avruch J, Chen L, Zhou D. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat Immunol. 2015; 16:1142–1152.
Article78. Wi SM, Moon G, Kim J, Kim ST, Shim JH, Chun E, Lee KY. TAK1-ECSIT-TRAF6 complex plays a key role in the TLR4 signal to activate NF-kappaB. J Biol Chem. 2014; 289:35205–35214.
Article79. Lei CQ, Zhang Y, Li M, Jiang LQ, Zhong B, Kim YH, Shu HB. ECSIT bridges RIG-I-like receptors to VISA in signaling events of innate antiviral responses. J Innate Immun. 2015; 7:153–164.
Article80. Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011; 469:221–225.
Article81. Khan M, Syed GH, Kim SJ, Siddiqui A. Hepatitis B Virus-Induced Parkin-Dependent Recruitment of Linear Ubiquitin Assembly Complex (LUBAC) to Mitochondria and Attenuation of Innate Immunity. PLoS Pathog. 2016; 12:e1005693.
Article82. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013; 501:512–516.
Article83. Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, Rentsendorj A, Vargas M, Guerrero C, Wang Y, Fitzgerald KA, Underhill DM, Town T, Arditi M. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012; 36:401–414.
Article84. Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, Matsuzawa S, Terskikh AV, Faustin B, Reed JC. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007; 129:45–56.
Article85. Bulua AC, Simon A, Maddipati R, Pelletier M, Park H, Kim KY, Sack MN, Kastner DL, Siegel RM. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J Exp Med. 2011; 208:519–533.
Article86. Kim HJ, Kim CH, Ryu JH, Kim MJ, Park CY, Lee JM, Holtzman MJ, Yoon JH. Reactive oxygen species induce antiviral innate immune response through IFN-lambda regulation in human nasal epithelial cells. Am J Respir Cell Mol Biol. 2013; 49:855–865.
Article87. Yang CS, Yuk JM, Kim JJ, Hwang JH, Lee CH, Kim JM, Oh GT, Choi HS, Jo EK. Small heterodimer partner-targeting therapy inhibits systemic inflammatory responses through mitochondrial uncoupling protein 2. PLoS One. 2013; 8:e63435.
Article88. Park J, Min JS, Kim B, Chae UB, Yun JW, Choi MS, Kong IK, Chang KT, Lee DS. Mitochondrial ROS govern the LPS-induced pro-inflammatory response in microglia cells by regulating MAPK and NF-kappaB pathways. Neurosci Lett. 2015; 584:191–196.
Article89. Mills EL, Kelly B, Logan A, Costa AS, Varma M, Bryant CE, Tourlomousis P, Dabritz JH, Gottlieb E, Latorre I, Corr SC, McManus G, Ryan D, Jacobs HT, Szibor M, Xavier RJ, Braun T, Frezza C, Murphy MP, O'Neill LA. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016; 167:457–470.
Article90. Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012; 485:251–255.
Article91. Sun S, Sursal T, Adibnia Y, Zhao C, Zheng Y, Li H, Otterbein LE, Hauser CJ, Itagaki K. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One. 2013; 8:e59989.
Article92. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015; 520:553–557.
Article93. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011; 12:222–230.
Article94. Won JH, Park S, Hong S, Son S, Yu JW. Rotenone-induced impairment of mitochondrial electron transport chain confers a selective priming signal for NLRP3 Inflammasome Activation. J Biol Chem. 2015; 290:27425–27437.
Article95. Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013; 14:454–460.
Article96. Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol. 2010; 299:C203–C210.
Article97. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012; 13:255–263.
Article98. Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: critical interplay between the two homeostats. Biochim Biophys Acta. 2012; 1820:595–600.
Article99. Rodgers MA, Bowman JW, Liang Q, Jung JU. Regulation where autophagy intersects the inflammasome. Antioxid Redox Signal. 2014; 20:495–506.
Article100. Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009; 106:2770–2775.
Article101. O'Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015; 43:331–342.102. Wang S, Xia P, Huang G, Zhu P, Liu J, Ye B, Du Y, Fan Z. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nat Commun. 2016; 7:11023.
Article