J Bacteriol Virol.
2013 Jun;43(2):111-119. 10.4167/jbv.2013.43.2.111.
Molecular Typing of Cryptococcus neoformans Isolated from Korean Patients
- Affiliations
-
- 1Department of Public Health, The Graduate School of Public Health, Seoul National University, Seoul, Korea.
- 2Department of Clinical Laboratory Science, Catholic University of Pusan, Busan, Korea. smhwang@cup.ac.kr
- KMID: 2168680
- DOI: http://doi.org/10.4167/jbv.2013.43.2.111
Abstract
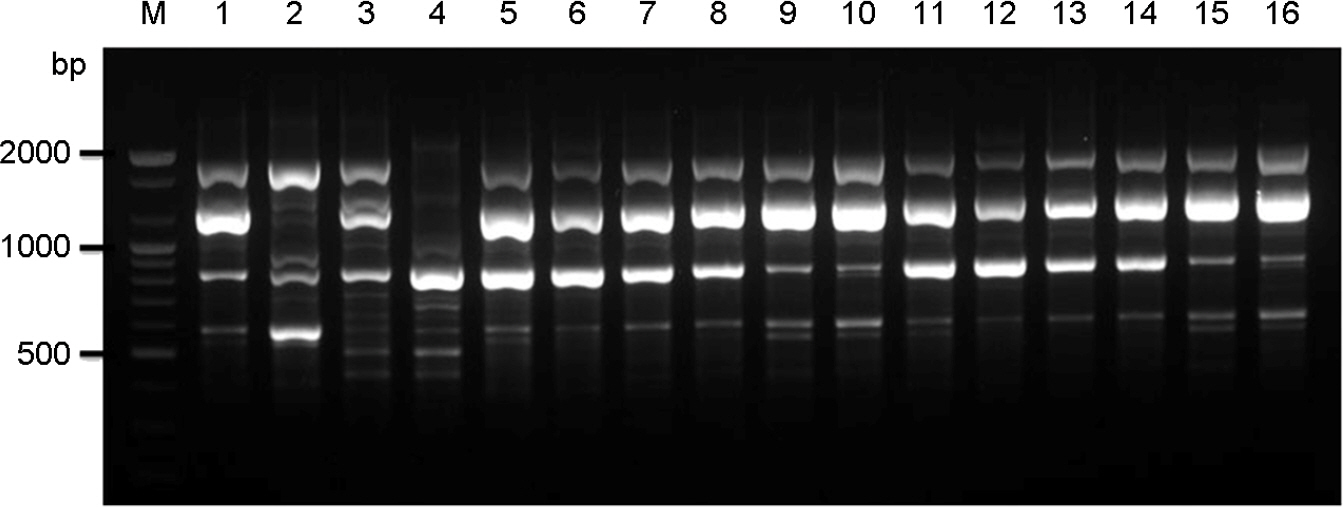
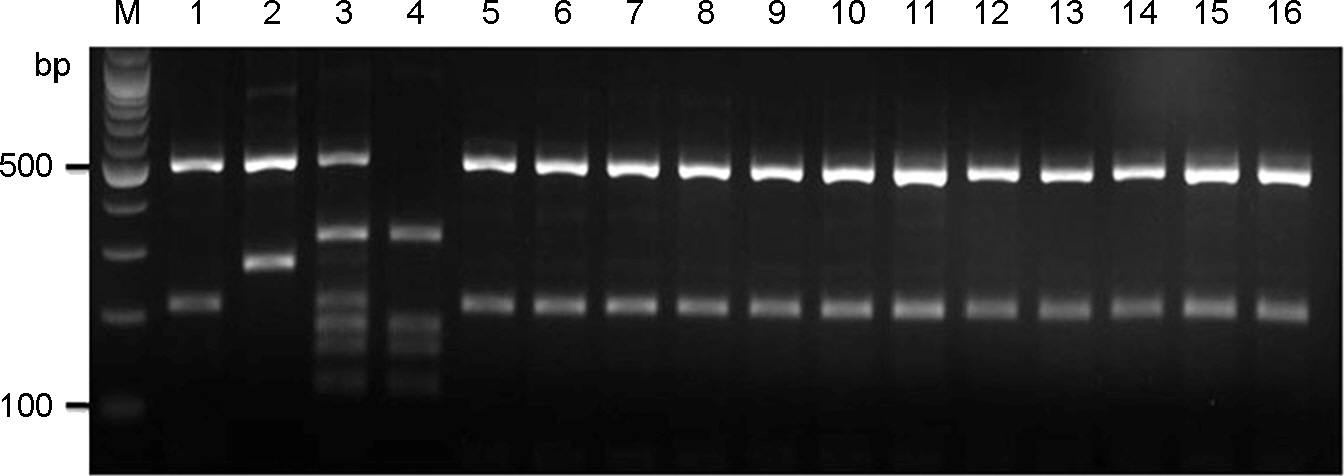
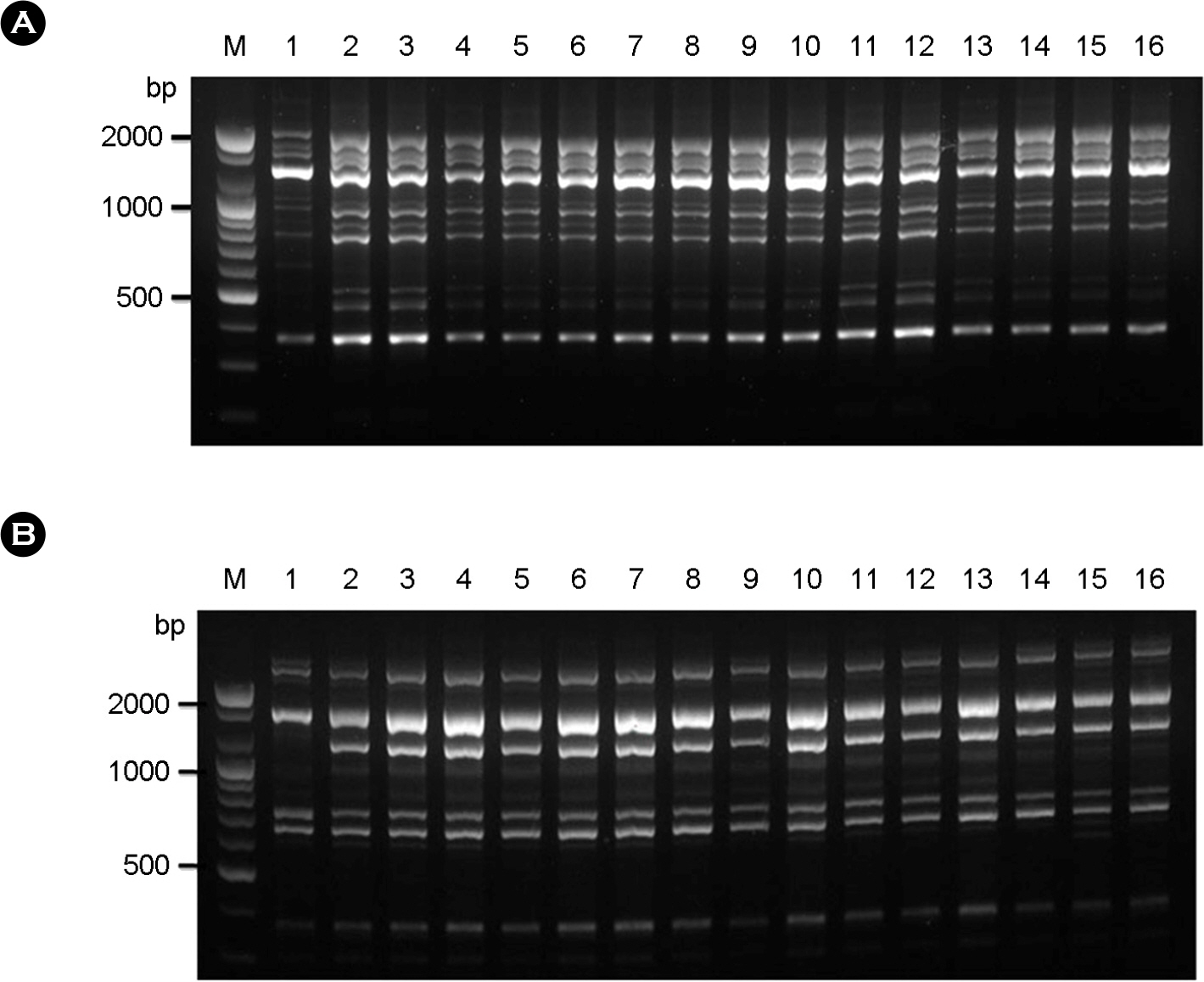
- Cyptococcosis is generally caused by Cryptococcus neoformans, the opportunistic agent which has two species such as C. neoformans and C. gattii. Both C. neoformans and C. gattii species contain a number of genetically diverse subgroups that can be differentiated by various molecular typing methods. We conducted a molecular epidemiological analysis of 30 clinical isolates of the C. neoformans from cryptococcosis patients who had been hospitalized between 2008 and 2010 in medical centers located in Seoul and Busan in Korea. To determine the genetic diversity, 30 strains of C. neoformans were typed using PCR fingerprinting with the microsatellite specific primer of the phage M13 and the restriction fragment length polymorphism (RFLP) of orotidine monophosphosphate pyrophosphorylase (URA5) gene. All isolates were identified as serotype A, mating type MATa and molecular type VNI. The random amplified polymorphic DNA (RAPD) profiles obtained by using two primers revealed a single pattern. Our study shows that 30 strains of clinical C. neoformans are genetically homogeneous, with all of the isolates were molecular type VN1, serotype A, mating type MATa.
MeSH Terms
Figure
Reference
-
1). Heiman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. Cryptococcus from human pathogen to model yeast. Washington D. C. USA: ASM Press;2011.2). Matsumoto MT, Fusco-Almeida AM, Baeza LC, Melhem Mde S, Medes-Giannini MJ. Genotyping, serotyping and determination of mating-type of Cryptococcus neoformans clinical isolates from São Paulo State, Brazil. Rev Inst Med Trop Sao Paulo. 2007; 49:41–7.3). Liaw SJ, Wu HC, Hsueh PR. Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clin Microbiol Infect. 2010; 16:696–703.4). Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008; 14:755–62.5). Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E. Molecular typing of Ibero American Cryptococcus neoformans isolates. Emerg Infect Dis. 2003; 9:189–95.6). Bennett JE, Kwon-Chung KJ, Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977; 105:582–6.7). Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990; 28:1642–4.8). Kuroki M, Phichaichumpon C, Yasuoka A, Chiranairadul P, Chosa T, Sirinirund P, et al. Environmental isolation of Cryptococcus neoformans from an endemic region of HIV-associated cryptococcosis in Thailand. Yeast. 2004; 21:809–12.9). Pappalardo MC, Melhem MS. Cryptococcosis: a review of the Brazilian experience for the disease. Rev Inst Med Trop Sao Paulo. 2003; 45:299–305.
Article10). Datta K, Bartlett KH, Marr KA. Cryptococcus gattii: emergence in western North America: exploitation of a novel ecological niche. Interdiscip Perspect Infect Dis. 2009; 2009:176532.11). Byrnes EJ 3rd, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, et al. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 2011; 7:e1002205.
Article12). Feng X, Yao Z, Ren D, Liao W, Wu J. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 2008; 8:930–8.13). Yan Z, Li X, Xu J. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J Clin Microbiol. 2002; 40:965–72.14). Chaturvedi S, Rodeghier B, Fan J, McClelland CM, Wickes BL, Chaturvedi V. Direct PCR of Cryptococcus neoformans MATalpha and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies. J Clin Microbiol. 2000; 38:2007–9.15). Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999; 20:1790–9.16). Staib F. The green colour effect (GCE) of the killer strain Cryptococcus laurentii CBS 139 on Staib agar. Mycoses. 1999; 42:103–6.17). Kwon-Chung KJ, Polacheck I, Bennett JE. Improved Diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotype B and C). J Clin Microbiol. 1982; 15:535–7.18). Yamamoto Y, Kohno S, Koga H, Kakeya H, Tomono K, Kaku M, et al. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J Clin Microbiol. 1995; 33:3328–32.19). Hwang SM. Molecular characterization of clinical and environmental strains of Cryptococcus neoformans isolated from Busan, Korea. J Bacteriol Virol. 2010; 40:91–8.20). Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006; 6:574–87.21). Illnait-Zaragozi MT, Martínez-Machín GF, Ferníndez-Andreu CM, Boekhout T, Meis JF, Klaassen CH. Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PLoS One. 2010; 5:e9124.22). Meyer W. Trilles, L. Genotyping of the Cryptococcus neoformans/C. gattii species complex. Australian Biochemist. 2010; 41:12–5.23). Ohkusu M, Tangonan N, Takeo K, Kishida E, Ohkubo M, Aoki S, et al. Serotype, mating type and ploidy of Cryptococcus neoformans strains isolated from patients in Brazil. Rev Inst Med Trop Sao Paulo. 2002; 44:299–302.24). Hwang SM, Oh KS, Lee KW. Serotype and enzymatic profile of Cryptococcus neoformans isolates from clinical and environmental sources in Korea. Korean J Microbiol. 2006; 42:257–64.25). Choi YH, Ngamskulrungroj P, Varma A, Sionov E, Hwang SM, Carriconde F, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 2010; 10:769–78.26). Hwang SM. Molecular typing of clinical Cryptococcus gattii isolates in Korea. J Bacteriol Virol. 2012; 42:152–5.27). Mihara T, Izumikawa K, Kakeya H, Ngamskulrungroj P, Umeyama T, Takazono T, et al. Multilocus sequence typing of Cryptococcus neoformans in non-HIV associated cryptococcosis in Nagasaki, Japan. Med Mycol. 2013; 51:252–60.28). Kwon-Chung KJ, Bennett JE. Distribution of α and α mating types of Cryptococcus neoformans among natural and clinical isolates. Am J Epidemiol. 1978; 108:337–40.29). Diaz MR, Boekhout T, Kiesling T, Fell JW. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Res. 2005; 5:1129–40.30). Brandt ME, Hutwagner LC, Kuykendall RJ, Pinner RW. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. The Cryplococcal Disease Active Surveillance Group. J Clin Microbiol. 1995; 33:1890–5.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genotypes of Clinical and Environmental Isolates of Cryptococcus neoformans and Cryptococcus gattii in Korea
- Differentiation of Varieties and Susceptibility Testing for Two Strains of Cryptococcus neoformans
- In vitro Evaluation of the Antifungal Activity of Propolis Extract on Cryptococcus neoformans and Candida albicans
- Isolation of Cryptococcus neoformans var. grubii (serotype A) from Pigeon Droppings in Korea
- Molecular Characterization of Clinical and Environmental Strains of Cryptococcus neoformans Isolated from Busan, Korea