J Periodontal Implant Sci.
2010 Feb;40(1):39-44. 10.5051/jpis.2010.40.1.39.
The synergistic regulatory effect of Runx2 and MEF transcription factors on osteoblast differentiation markers
- Affiliations
-
- 1Department of Periodontology, Kyungpook National University School of Dentistry, Daegu, Korea.
- 2BIDMC Genomics Center, Harvard Medical School, Boston, USA.
- 3Department of Oral Biochemistry, Kyungpook National University School of Dentistry, Daegu, Korea. jeycho@knu.ac.kr
- KMID: 2094714
- DOI: http://doi.org/10.5051/jpis.2010.40.1.39
Abstract
- PURPOSE
Bone tissues for clinical application can be improved by studies on osteoblast differentiation. Runx2 is known to be an important transcription factor for osteoblast differentiation. However, bone morphogenetic protein (BMP)-2 treatment to stimulate Runx2 is not sufficient to acquire enough bone formation in osteoblasts. Therefore, it is necessary to find other regulatory factors which can improve the transcriptional activity of Runx2. The erythroblast transformation-specific (ETS) transcription factor family is reported to be involved in various aspects of cellular proliferation and differentiation.
METHODS
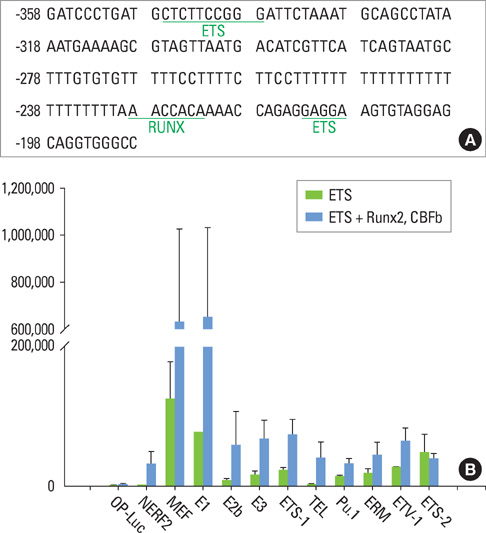
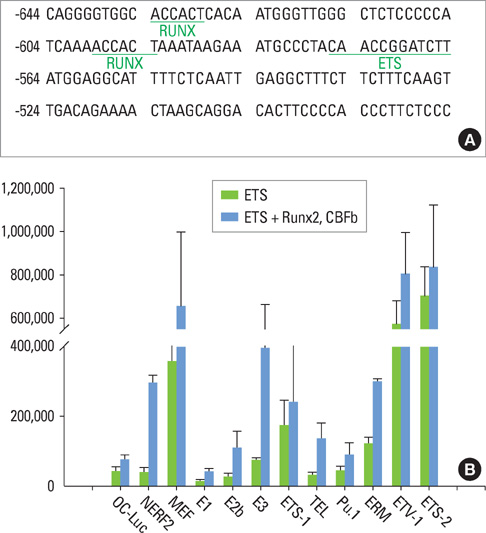
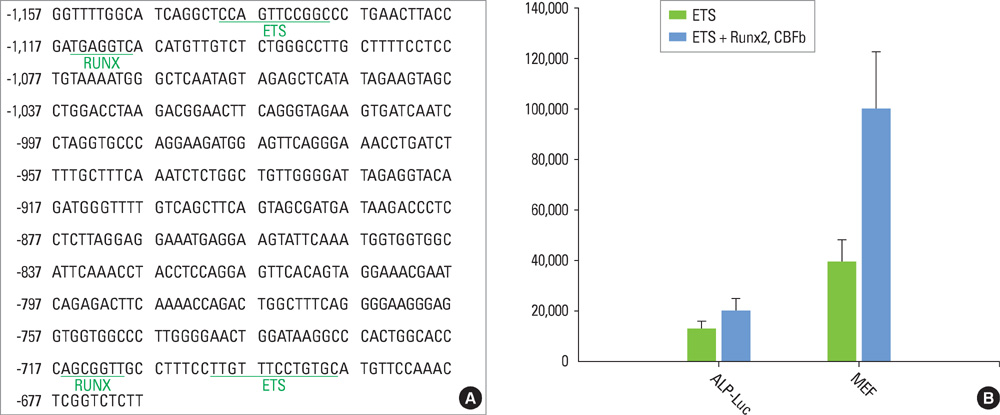
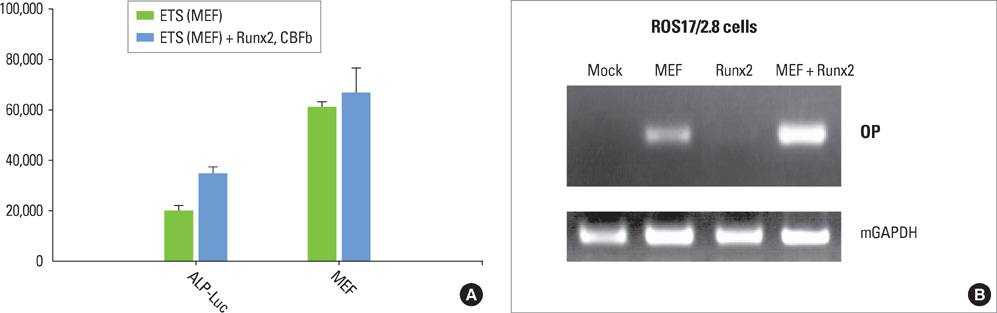
We have noticed that the promoters of osteoblast differentiation markers such as alkaline phosphatase (Alp), osteopontin (Opn), and osteocalcin (Oc) contain Ets binding sequences which are also close to Runx2 binding elements. Luciferase assays were performed to measure the promoter activities of these osteoblast differentiation markers after the transfection of Runx2, myeloid Elf-1-like factor (MEF), and Runxs+MEF. Reverse-transcription polymerase chain reaction was also done to check the mRNA levels of Opn after Runx2 and MEF transfection into rat osteoblast (ROS) cells.
RESULTS
We have found that MEF, an Ets transcription factor, increased the transcriptional activities of Alp, Opn, and Oc. The addition of Runx2 resulted in the 2- to 6-fold increase of the activities. This means that these two transcription factors have a synergistic effect on the osteoblast differentiation markers. Furthermore, early introduction of these two Runx2 and MEF factors significantly elevated the expression of the Opn mRNA levels in ROS cells. We also showed that Runx2 and MEF proteins physically interact with each other.
CONCLUSIONS
Runx2 interacts with MEF proteins and binds to the promoters of the osteoblast markers such as Opn nearby MEF to increase its transcriptional activity. Our results also imply that osteoblast differentiation and bone formation can be increased by activating MEF to elicit the synergistic effect of Runx2 and MEF.
MeSH Terms
-
Alkaline Phosphatase
Animals
Antigens, Differentiation
Bone and Bones
Bone Morphogenetic Proteins
Cell Differentiation
Cell Proliferation
Core Binding Factor Alpha 1 Subunit
Erythroblasts
Humans
Luciferases
Osteoblasts
Osteocalcin
Osteogenesis
Osteopontin
Polymerase Chain Reaction
Proteins
Rats
RNA, Messenger
Transcription Factors
Transfection
Alkaline Phosphatase
Antigens, Differentiation
Bone Morphogenetic Proteins
Core Binding Factor Alpha 1 Subunit
Luciferases
Osteocalcin
Osteopontin
Proteins
RNA, Messenger
Transcription Factors
Figure
Reference
-
1. Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003. 423:337–342.
Article2. Blair HC, Zaidi M, Schlesinger PH. Mechanisms balancing skeletal matrix synthesis and degradation. Biochem J. 2002. 364:329–341.
Article3. Teitelbaum SL. Osteoclasts, integrins, and osteoporosis. J Bone Mineral Metab. 2000. 18:344–349.
Article4. Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002. 2:389–406.
Article5. Lemaire V, Tobin FL, Greller LD, Cho CR, Suva LJ. Modeling the interactions between osteoblast and osteoclast activities in bone remodeling. J Theor Biol. 2004. 229:293–309.
Article6. Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000. 289:1504–1508.
Article7. Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000. 289:1501–1504.
Article8. Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003. 423:349–355.
Article9. Olsen BR, Reginato AM, Wang W. Bone development. Ann Rev Cell Dev Biol. 2000. 16:191–220.
Article10. Katagiri T, Takahashi N. Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis. 2002. 8:147–159.
Article11. Wagner EF, Karsenty G. Genetic control of skeletal development. Curr Opin Genet Dev. 2001. 11:527–532.
Article12. Deng ZL, Sharff KA, Tang N, Song WX, Luo J, Luo X, et al. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 2008. 13:2001–2021.
Article13. Raouf A, Seth A. Ets transcription factors and targets in osteogenesis. Oncogene. 2000. 19:6455–6463.
Article14. Miyazaki Y, Sun X, Uchida H, Zhang J, Nimer S. MEF, a novel transcription factor with an Elf-1 like DNA binding domain but distinct transcriptional activating properties. Oncogene. 1996. 13:1721–1729.15. Suico MA, Koyanagi T, Ise S, Lu Z, Hisatsune A, Seki Y, et al. Functional dissection of the ETS transcription factor MEF. Biochim Biophys Acta. 2002. 1577:113–120.
Article16. Hedvat CV, Yao J, Sokolic RA, Nimer SD. Myeloid ELF-1 like factor is a potent activator of interleukin-8 expression in hematopoietic cells. J Biol Chem. 2004. 279:6395–6400.
Article17. Lu Z, Kim KA, Suico MA, Shuto T, Li JD, Kai H. MEF up-regulates human beta-defensin 2 expression in epithelial cells. FEBS Lett. 2004. 561:117–121.18. Lee MH, Kim YJ, Kim HJ, Park HD, Kang AR, Kyung HM, et al. BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-beta 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J Biol Chem. 2003. 278:34387–34394.
Article19. Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, Yamaguchi A, et al. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem. 1999. 274:6972–6978.
Article20. Towler DA, Bennett CD, Rodan GA. Activity of the rat osteocalcin basal promoter in osteoblastic cells is dependent upon homeodomain and CP1 binding motifs. Mol Endocrinol. 1994. 8:614–624.
Article21. Towler DA, Rutledge SJ, Rodan GA. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol Endocrinol. 1994. 8:1484–1493.
Article22. Lacorazza HD, Nimer SD. The emerging role of the myeloid Elf-1 like transcription factor in hematopoiesis. Blood Cells Mol Dis. 2003. 31:342–350.
Article23. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997. 89:755–764.
Article24. Sato M, Morii E, Komori T, Kawahata H, Sugimoto M, Terai K, et al. Transcriptional regulation of osteopontin gene in vivo by PEBP2alphaA/CBFA1 and ETS1 in the skeletal tissues. Oncogene. 1998. 17:1517–1525.
Article25. Wai PY, Mi Z, Gao C, Guo H, Marroquin C, Kuo PC. Ets-1 and runx2 regulate transcription of a metastatic gene, osteopontin, in murine colorectal cancer cells. J Biol Chem. 2006. 281:18973–18982.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Function of runx2 and osterix in osteogenesis and teeth
- Zinc upregulates bone-specific transcription factor Runx2 expression via BMP-2 signaling and Smad-1 phosphorylation in osteoblasts
- Aqueous extract of Petasites japonicus leaves promotes osteoblast differentiation via up-regulation of Runx2 and Osterix in MC3T3-E1 cells
- Synergistic Effects of Chios Gum Mastic Extract and Low Level Laser Therapy on Osteoblast Differentiation
- Immunolocalization of Runx2 and Osterix in the Developing Periodontal Tissues of the Mouse