Infect Chemother.
2008 Jun;40(3):167-169. 10.3947/ic.2008.40.3.167.
Comparison of Real-time PCR Methods and pp65 Antigenemia Assay to Detect Cytomegalovirus Reactivation in Hematopoietic Stem Cell Transplantation
- Affiliations
-
- 1Center for Immunology and Pathology, National Institute of Health, Seoul, Korea. jooshil@nih.go.kr
- 2The Catholic Hematopoietic Stem Cell Transplantation Center, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 1782290
- DOI: http://doi.org/10.3947/ic.2008.40.3.167
Abstract
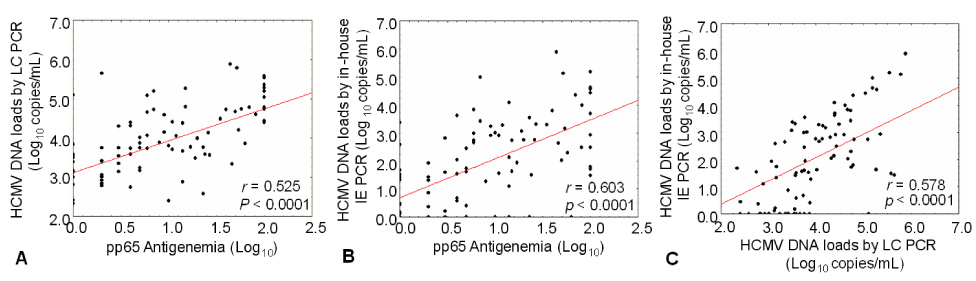
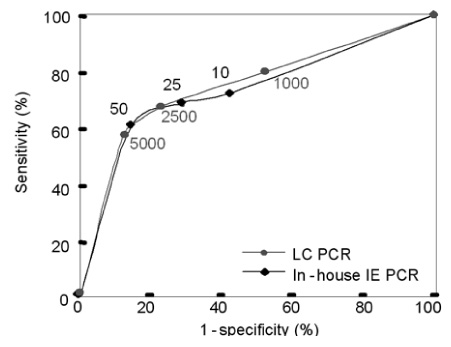
- Human cytomegalovirus (HCMV) is a common human pathogen that causes morbidity and mortality in hematopoietic stem cell transplantation (HSCT) recipients. Early diagnosis of HCMV infection or reactivation, and setting threshold values for effective pre-emptive therapies, are required for appropriate HCMV disease prevention in HSCT recipients. We compared the HCMV infections detected by the two methods, LightCycler-based PCR (LC PCR) and in-house immediate early protein PCR (in-house IE PCR) with the results of a pp65 antigenemia assay as the reference. The sensitivity and specificity for the in-house IE PCR were 79.3% and 72.7%, respectively, and 82.9% and 40.7%, respectively, for the LC PCR. The correlation between the HCMV viral load and pp65 antigenemia in HSCT recipients was r=0.603 with in-house IE PCR and r=0.525 with LC PCR. The discordant results between methods and relatively low (r) values suggest that we need more study to set threshold values according to the using methods with clinical outcome.
Keyword
MeSH Terms
Figure
Reference
-
1. Allice T, Enrietto M, Pittaluga F, Varetto S, Franchello A, Marchiaro G, Ghisetti V. Quantitation of cytomegalovirus DNA by real-time polymerase chain reaction in peripheral blood specimens of patients with solid organ transplants: comparison with endpoint PCR and pp65 antigen test. J Med Virol. 2006. 78:915–922.
Article2. Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L. Optimization of Quantitative Detection of Cytomegalovirus DNA in Plasma by Real-Time PCR. J Clin Microbiol. 2004. 42:1142–1148.
Article3. Pumannova M, Roubalova K, Vitek A, Sajdova J. Comparison of quantitative competitive polymerase chain reaction-enzyme-linked immunosorbent assay with LightCycler-based polymerase chain reaction for measuring cytomegalovirus DNA in patients after hematopoietic stem cell transplantation. Diagn Microbiol Infect Dis. 2006. 54:115–120.
Article4. Boeckh M, Gallez-Hawkins GM, Myerson D, Zaia JA, Bowden RA. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation. 1997. 64:108–113.
Article5. Gor D, Sabin C, Prentice HG, Vyas N, Man S, Griffiths PD, Emery VC. Longitudinal fluctuations in cytomegalovirus load in bone marrow transplant patients: relationship between peak virus load, donor/ recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transplant. 1998. 21:597–605.
Article6. Ikewaki J, Ohtsuka E, Kawano R, Ogata M, Kikuchi H, Nasu M. Real-time PCR assay compared to nested PCR and antigenemia assays for detecting cytomegalovirus reactivation in adult T-cell leukemialymphoma patients. J Clin Microbiol. 2003. 41:4382–4387.
Article7. Ikewaki J, Ohtsuka E, Satou T, Kawano R, Ogata M, Kikuchi H, Nasu M. Real-time PCR assays based on distinct genomic regions for cytomegalovirus reactivation following hematopoietic stem cell transplantation. Bone Marrow Transplant. 2005. 35:403–410.
Article8. Mori T, Okamoto S, Watanabe R, Yajima T, Iwao Y, Yamazaki R, Nakazato T, Sato N, Iguchi T, Nagayama H, Takayama N, Hibi T, Ikeda Y. Dose-adjusted preemptive therapy for cytomegalovirus disease based on real-time polymerase chain reaction after allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 2002. 29:777–782.
Article9. Choi SM, Lee DG, Choi JH, Yoo JH, Kim YJ, Park SH, Park SN, Min CK, Lee S, Kim HJ, Kim DW, Lee JW, Min WS, Shin WS, Kim CC. Risk-adapted preemptive therapy for cytomegalovirus disease after allogeneic stem cell transplantation: a single-center experience in Korea. Int J Hematol. 2005. 81:69–74.
Article10. Yakushiji K, Gondo H, Kamezaki K, Shigematsu K, Hayashi S, Kuroiwa M, Taniguchi S, Ohno Y, Takase K, Numata A, Aoki K, Kato K, Nagafuji K, Shimoda K, Okamura T, Kinukawa N, Kasuga N, Sata M, Harada M. Monitoring of cytomegalovirus reactivation after allogeneic stem cell transplantation: comparison of an antigenemia assay and quantitative real-time polymerase chain reaction. Bone Marrow Transplant. 2002. 29:599–606.
Article11. Li H, Dummer JS, Estes WR, Meng S, Wright PF, Tang YW. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J Clin Microbiol. 2003. 41:187–191.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparison of Quantitative Cytomegalovirus Real-time PCR in Whole Blood and pp65 Antigenemia Assay: Clinical Utility of CMV Real-time PCR in Hematopoietic Stem Cell Transplant Recipients
- Rapid Diagnosis of HCMV Diseases by pp65 Antigenemia Assay: Comparison of pp65 Antigenemia with Polymerase Chain Reaction
- Comparison of Quantitation of Cytomegalovirus DNA by Real-Time PCR in Whole Blood with the Cytomegalovirus Antigenemia Assay
- Evaluation of BiosewoomTM Real-Q Cytomegalovirus Quantification kit for Cytomegalovirus Viral Load Measure
- Primary Cytomegalovirus Peritonitis Following Unrelated Hematopoietic Stem Cell Transplantation