Korean J Lab Med.
2007 Aug;27(4):298-304. 10.3343/kjlm.2007.27.4.298.
Evaluation of BiosewoomTM Real-Q Cytomegalovirus Quantification kit for Cytomegalovirus Viral Load Measure
- Affiliations
-
- 1Department of Laboratory Medicine, Kyung Pook National University Hospital, Daegu, Korea. suhjs@knu.ac.kr
- 2Biosewoom Institute of Bioscience & Biotechnology, Seoul, Korea.
- 3Department of Laboratory Medicine, University of Ulsan College of Medicine and Asan Medical Center, Seoul, Korea.
- KMID: 1781500
- DOI: http://doi.org/10.3343/kjlm.2007.27.4.298
Abstract
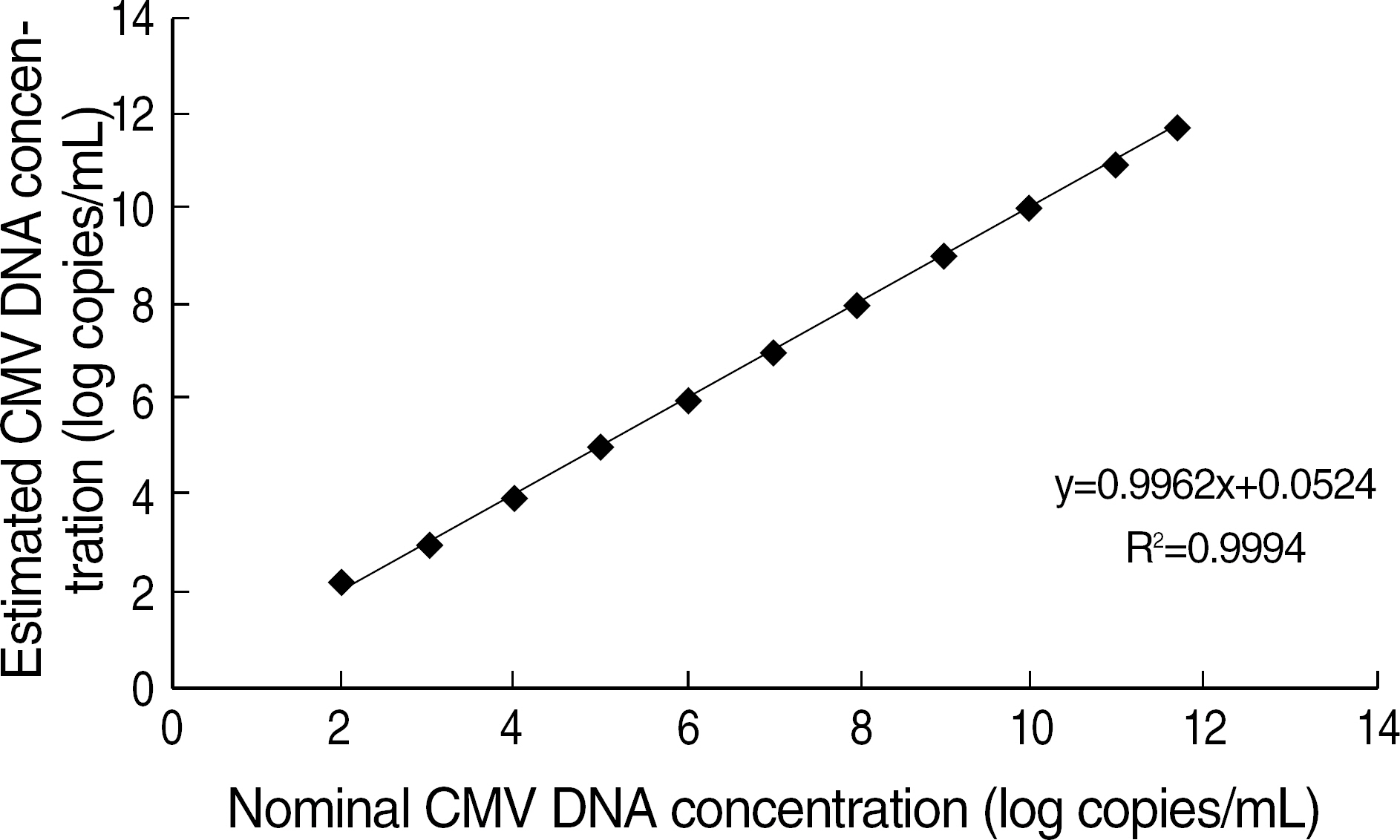
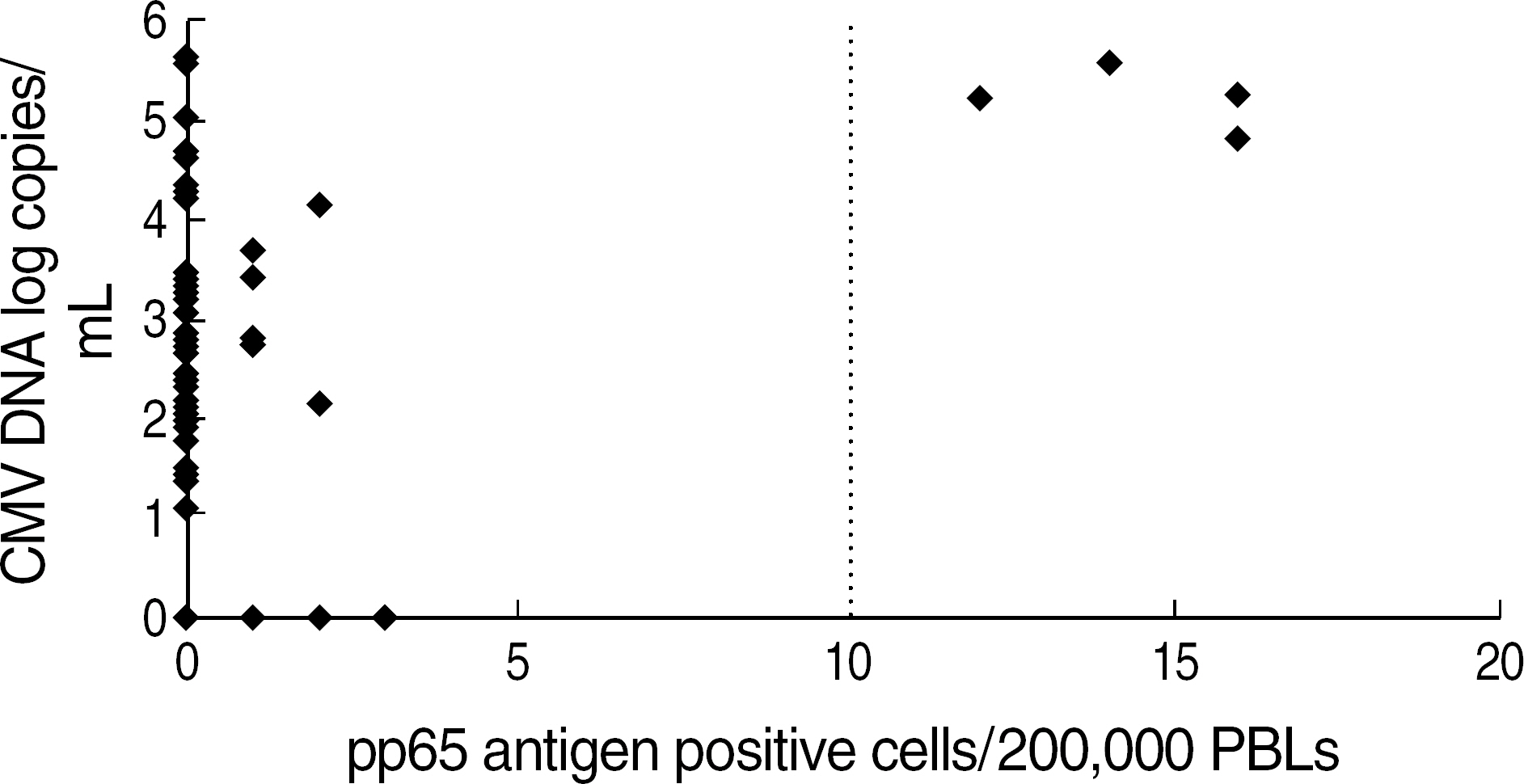
- BACKGROUND: Rapid and accurate laboratory tests are essential to detect cytomegalovirus (CMV) infections in solid organs and haematopoietic stem cell transplant recipients. We assessed the realtime quantitative PCR (RQ-PCR) technology for its usefulness in detecting CMV DNA. METHODS: We evaluated the analytical performance of CMV RQ-PCR using Real-Q Cytomegalovirus Quantification kit (BioSewoom Inc., Korea). To evaluate its clinical utility, we also compared it to pp65 antigenemia test, an immunostaining method, on 343 samples of total 84 patients, including 63 transplant recipients. RESULTS: The detection limit of RQ-PCR was 63 copies/mL and none of hepatitis B virus, hepatitis C virus, or human immunodeficiency virus showed a cross-reactivity with CMV. Total coefficient of variation (CV) was 10.4-19.5%. It detected CMV DNA in a linear range from 1 x 10(2) to 5 x 10(11) copies/mL (P<10(-13), R2=0.9994). The qualitative positive rates of pp65 antigenemia test and RQ-PCR were 4.7%, 16.3%, respectively and concordance rate between the two tests was 84.8% (K=0.221, P<10(-6)). In comparison of quantitative results, the correlation between two tests was significant (r=0.45, P<10(-17)). In comparison among three groups by pp65 antigen level, CMV DNA level obtained with RQ-PCR increased significantly (P<10(-3) and P<10(-7), respectively). CONCLUSIONS: The RQ-PCR is easier to perform than the immunostaining method, has good analytical performance and reflects the blood level of viral DNA well. It may be a new method substituting the pp65 antigenemia test. Further studies determining RQ-PCR value starting pre-emptive therapy will be required.
Keyword
MeSH Terms
-
Adolescent
Adult
Aged
Child
Child, Preschool
Cytomegalovirus/genetics/*isolation & purification
Cytomegalovirus Infections/*diagnosis/virology
DNA, Viral/blood
Female
Humans
Infant
Infant, Newborn
Male
Middle Aged
Phosphoproteins/analysis
Polymerase Chain Reaction/*methods
Reagent Kits, Diagnostic
Reproducibility of Results
Sensitivity and Specificity
Viral Load/*methods
Viral Matrix Proteins/analysis/blood
Figure
Reference
-
References
1. Paya CV. Prevention of cytomegalovirus disease in recipients of solid-organ transplants. Clin Infect Dis. 2001; 32:596–603.2. Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998; 11:533–54.
Article3. Camargo LF, Uip DE, Simpson AA, Caballero O, Stolf NA, Vilas-Boas LS, et al. Comparison between antigenemia and a quantitative-competitive polymerase chain reaction for the diagnosis of cytomegalovirus infection after heart transplantation. Transplantation. 2001; 71:412–7.4. Guiver M, Fox AJ, Mutton K, Mogulkoc N, Egan J. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation. 2001; 71:1609–15.
Article5. Monte PD, Lazzarotto T, Ripalti A, Landini MP. Human cytomegalovirus infection: a complex diagnostic problem in which molecular biology has induced a rapid evolution. Intervirology. 1996; 39:193–203.
Article6. Delgado R, Lumbreras C, Alba C, Pedraza MA, Otero JR, Gomez R, et al. Low predictive value of polymerase chain reaction for diagnosis of cytomegalovirus disease in liver transplant recipients. J Clin Microbiol. 1992; 30:1876–8.
Article7. Schaade L, Kockelkorn P, Ritter K, Kleines M. Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J Clin Microbiol. 2000; 38:4006–9.
Article8. Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002; 34:1094–7.
Article9. Patel R, Snydman DR, Rubin RH, Ho M, Pescovitz M, Martin M, et al. Cytomegalovirus prophylaxis in solid organ transplant recipients. Transplantation. 1996; 61:1279–89.
Article10. Kearns AM, Guiver M, James V, King J. Development and evaluation of a real-time quantitative PCR for the detection of human cytomegalovirus. J Virol Methods. 2001; 95:121–31.
Article11. Mandell GL, Bennett JE, editors. Principles and practices of infectious diseases. 6th ed.Philadelphia: Elsevier Churchill Livingstone;2005. p. 1786–7.12. Meyers JD, Flournoy N, Thomas ED. Risk factors for cytomegalovirus infection after human marrow transplantation. J Infect Dis. 1986; 153:478–88.
Article13. Masur H, Whitcup SM, Cartwright C, Polis M, Nussenblatt R. Advances in the management of AIDS-related cytomegalovirus retinitis. Ann Intern Med. 1996; 125:126–36.
Article14. Nokta MA, Holland F, De Gruttola V, Emery VC, Jacobson MA, Griffiths P, et al. Cytomegalovirus (CMV) polymerase chain reaction profiles in individuals with advanced human immunodeficiency virus infection: relationship to CMV disease. J Infect Dis. 2002; 185:1717–22.
Article15. Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet. 2000; 355:2032–6.
Article16. Sia IG, Wilson JA, Espy MJ, Paya CV, Smith TF. Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J Clin Microbiol. 2000; 38:600–6.
Article17. Allice T, Enrietto M, Pittaluga F, Varetto S, Franchello A, Marchiaro G, et al. Quantitation of cytomegalovirus DNA by real-time polymerase chain reaction in peripheral blood specimens of patients with solid organ transplants: comparison with end-point PCR and pp65 antigen test. J Med Virol. 2006; 78:915–22.
Article18. Piiparinen H, Lautenschlager I. Quantitative TaqMan assay for the detection and monitoring of cytomegalovirus infection in organ transplant patients. Methods Mol Biol. 2006; 335:147–56.
Article19. Martin-Davila P, Fortun J, Gutierrez C, Marti-Belda P, Candelas A, Honrubia A, et al. Analysis of a quantitative PCR assay for CMV infection in liver transplant recipients: an intent to find the optimal cut-off value. J Clin Virol. 2005; 33:138–44.20. Hernando S, Folgueira L, Lumbreras C, San Juan R, Maldonado S, Prieto C, et al. Comparison of cytomegalovirus viral load measure by real-time PCR with pp65 antigenemia for the diagnosis of cytomegalovirus disease in solid organ transplant patients. Transplant Proc. 2005; 37:4094–6.
Article21. Fan J, Ma WH, Yang MF, Xue H, Gao HN, Li LJ. Real-time fluorescent quantitative PCR assay for measuring cytomegalovirus DNA load in patients after haematopoietic stem cell transplantation. Chin Med J. 2006; 119:871–4.
Article22. Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996; 70:78–83.
Article23. Zweygberg Wirgart B, Brytting M, Linde A, Wahren B, Grillner L. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J Clin Microbiol. 1998; 36:3662–9.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Performance Evaluation of the Real-Q Cytomegalovirus (CMV) Quantification Kit Using Two Real-Time PCR Systems for Quantifying CMV DNA in Whole Blood
- Comparison Cytomegalovirus Qualitative Assay Using Real-Time PCR and Conventional PCR
- Performance Evaluation of the ELITe InGenius System for Detecting Cytomegalovirus, EpsteinBarr Virus, and BK Virus Infections
- Comparison of Quantitation of Cytomegalovirus DNA by Real-Time PCR in Whole Blood with the Cytomegalovirus Antigenemia Assay
- Cytomegalovirus Infections in Solid Organ Transplantation: A Review