J Korean Med Sci.
2005 Aug;20(4):579-585. 10.3346/jkms.2005.20.4.579.
Proteomic Analysis of Differently Expressed Proteins in a Mouse Model for Allergic Asthma
- Affiliations
-
- 1Genome Research Center for Allergy and Respiratory Diseases, Soonchunhyang University Hospital, Korea. sung237@unitel.co.kr
- 2Department of Chemistry, Soonchunhyang University, Korea.
- 3Department of Biochemistry and Molecular Biology, Hanyang University, Seoul, Korea.
- KMID: 1712736
- DOI: http://doi.org/10.3346/jkms.2005.20.4.579
Abstract
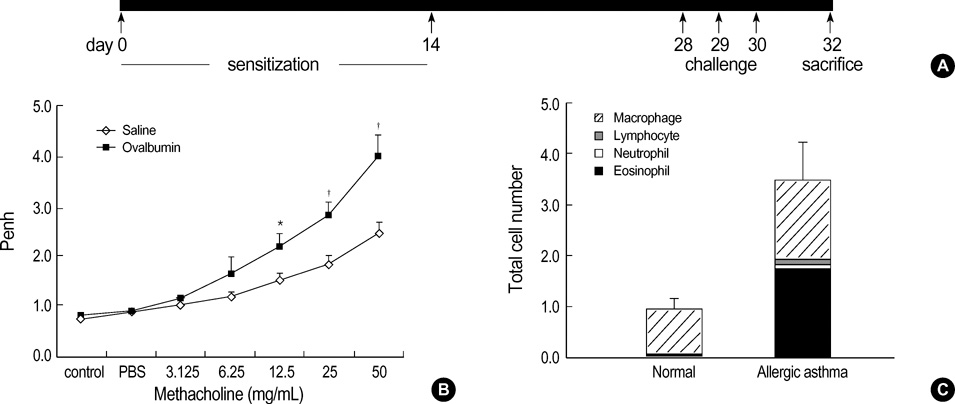
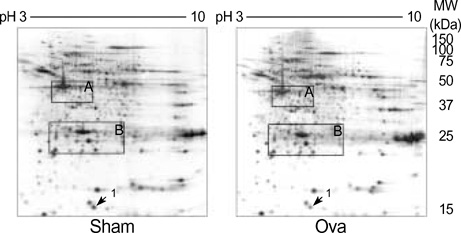
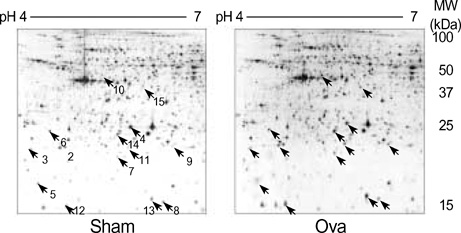
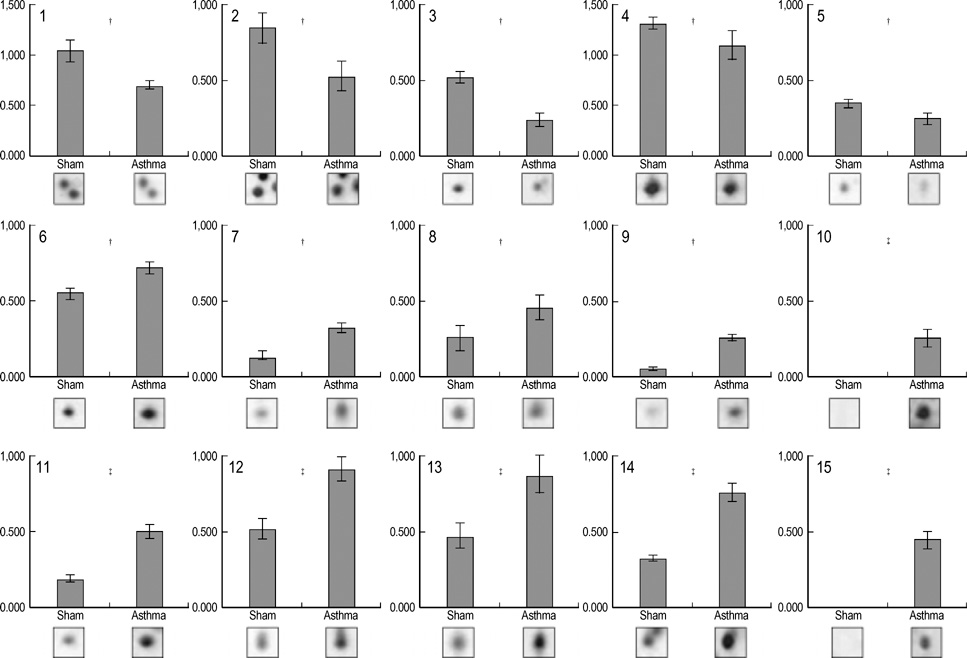
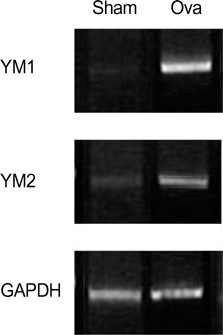
- Allergic asthma is associated with persistent functional and structural changes in the airways and involves many different cell types. Many proteins involved in allergic asthma have been identified individually, but complete protein profiles (proteome) have not yet been reported. Here we have used a differential proteome mapping strategy to identify tissue proteins that are differentially expressed in mice with allergic asthma and in normal mice. Mouse lung tissue proteins were separated using two-dimensional gel electrophoresis over a pH range between 4 and 7, digested, and then analyzed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MS). The proteins were identified using automated MS data acquisition. The resulting data were searched against a protein database using an internal Mascot search routine. This approach identified 15 proteins that were differentially expressed in the lungs of mice with allergic asthma and normal mice. All 15 proteins were identified by MS, and 9 could be linked to asthma-related symptoms, oxidation, or tissue remodeling. Our data suggest that these proteins may prove useful as surrogate biomarkers for quantitatively monitoring disease state progression or response to therapy.
Keyword
MeSH Terms
-
Animals
Asthma/genetics/immunology/*metabolism
Comparative Study
Disease Models, Animal
Electrophoresis, Gel, Two-Dimensional
Gene Expression/immunology
Gene Expression Profiling
Lung/immunology/metabolism/pathology
Male
Mice
Mice, Inbred BALB C
Ovalbumin/immunology
Proteome/*analysis/genetics/immunology
Proteomics/methods
RNA, Messenger/genetics/metabolism
Research Support, Non-U.S. Gov't
Reverse Transcriptase Polymerase Chain Reaction
Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization
Figure
Reference
-
1. Jember AG, Zuberi R, Liu FT, Croft M. Development of allergic inflammation in a murine model of asthma is dependent on the costimulatory receptor OX40. J Exp Med. 2001. 193:387–392.
Article2. Hakonarson H, Halapi E. Genetic analyses in asthma: current concepts and future directions. Am J Pharmacogenomics. 2002. 2:155–166.3. Busse WW, Lemanske RF Jr. Asthma. N Engl J Med. 2001. 344:350–362.
Article4. Hakonarson H, Bjornsdottir US, Halapi E, Palsson S, Adalsteinsdottir E, Gislason D, Finnbogason G, Gislason T, Kristjansson K, Arnason T, Birkisson I, Frigge ML, Kong A, Gulcher JR, Stefansson K. A major susceptibility gene for asthma maps to chromosome 14q24. Am J Hum Genet. 2002. 71:483–491.
Article5. Coyle AJ, Le Gros G, Bertrand C, Tsuyuki S, Heusser CH, Kopf M, Anderson GP. Interleukin-4 is required for the induction of lung Th2 mucosal immunity. Am J Respir Cell Mol Biol. 1995. 13:54–59.
Article6. Foster PS, Hogan SP, Ramsay AJ, Matthaei KI, Young IG. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J Exp Med. 1996. 183:195–201.
Article7. Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, Sheppard D, Mohrs M, Donaldson DD, Locksley RM, Corry DB. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998. 282:2261–2263.
Article8. Cohn L, Homer RJ, MacLeod H, Mohrs M, Brombacher F, Bottomly K. Th2-induced airway mucus production is dependent on IL-4Ralpha, but not on eosinophils. J Immunol. 1999. 162:6178–6183.9. Townsend MJ, Fallon PG, Matthews DJ, Smith P, Jolin HE, McKenzie AN. IL-9-deficient mice establish fundamental roles for IL-9 in pulmonary mastocytosis and goblet cell hyperplasia but not T cell development. Immunity. 2000. 13:573–583.
Article10. Gavett SH, Chen X, Finkelman F, Wills-Karp M. Depletion of murine CD4+ T lymphocytes prevents antigen-induced airway hyperreactivity and pulmonary eosinophilia. Am J Respir Cell Mol Biol. 1994. 10:587–593.11. O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975. 250:4007–4021.12. Gorg A, Postel W, Gunther S. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988. 9:531–546.13. Rabilloud T, Adessi C, Giraudel A, Lunardi J. Improvement of the solubilization of proteins in two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1997. 18:307–316.
Article14. Molloy MP, Herbert BR, Walsh BJ, Tyler MI, Traini M, Sanchez JC, Hochstrasser DF, Williams KL, Gooley AA. Extraction of membrane proteins by differential solubilization for separation using twodimensional gel electrophoresis. Electrophoresis. 1998. 19:837–844.
Article15. Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: Large-scale identification of yeast proteins from two-dimensional gels. Proc Natl Acad Sci USA. 1996. 93:14440–14445.16. Dainese P, Staudenmann W, Quadroni M, Korostensky C, Gonnet G, Kertesz M, James P. Probing protein function using a combination of gene knockout and proteome analysis by mass spectrometry. Electrophoresis. 1997. 18:432–442.
Article17. Roepstorff P. Mass spectrometry in protein studies: From genome to function. Curr Opin Biotechnol. 1997. 8:6–13.
Article18. Yates JR III. Mass spectrometry and the age of the proteome. J Mass Spectrom. 1998. 33:1–19.
Article19. Hamelmann E, Schwarze J, Takeda K, Oshiba A, Larsen GL, Irvin CG, Gelfand EW. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Am J Respir Crit Care Med. 1997. 156:766–775.
Article20. Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, McNulty DE, Carr SA, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998. 826:167–181.
Article21. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996. 68:850–858.22. Beavis RC, Chait BT. a-Cyano-4-hydroxycinnamic Acid as a Matrix for Matrix assisted Laser Desorption mass Spectrometry. Org Mass Spectrom. 1992. 27:156–158.23. Vorm O, Roepstorff P, Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994. 66:3281–3287.
Article24. Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem. 2002. 383:347–364.
Article25. Hegesh E, Hegesh J, Kaftory A. Congenital methemoglobinemia with a deficiency of cytochrome b5. N Engl J Med. 1986. 314:757–761.
Article26. Hultquist DE, Passon PG. Catalysis of methaemoglobin reduction by erythrocyte cytochrome b5 and cytochrome b5 reductase. Nature New Biol. 1971. 229:252–254.
Article27. Fish JE, Peters SP. Airway remodeling and persistent airway obstruction in asthma J. Allergy Clin Immunol. 1999. 104(3 Pt 1):509–516.28. Adra CN, Ko J, Leonard D, Wirth LJ, Cerione RA, Lim B. Identification of a novel protein with GDP dissociation inhibitor activity for the ras-like proteins CDC42Hs and rac I. Genes Chromosomes Cancer. 1993. 8:253–261.29. Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass CK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem. 2002. 277:42821–42829.
Article30. Webb DC, McKenzie AN, Foster PS. Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J Biol Chem. 2001. 276:41969–41976.31. Zhu Z, Zheng T, Homer RJ, Kim Y, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004. 304:1678–1682.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Allergic March: Progression from Atopic Dermatitis to Asthma
- Principles and Application of Mouse Model of Allergic Rhinitis
- Understanding the Mouse Model of Respiratory Allergic Diseases
- Allergic Rhinitis Mouse Model
- Immunological pathogenesis of asthma in a mouse model Can asthma pathogenesis be explained by allergic reaction?