Cancer Res Treat.
2023 Oct;55(4):1270-1280. 10.4143/crt.2023.415.
Specific Mutations in APC, with Prognostic Implications in Metastatic Colorectal Cancer
- Affiliations
-
- 1Division of Colorectal Surgery, Department of Surgery, Second Affiliated Hospital of Naval Medical University, Shanghai, China
- 2Jiaxing Key Laboratory of Precision Medicine and Companion Diagnostics, Jiaxing Yunying Medical Inspection Co., Ltd., Jiaxing, China
- 3Department of R&D, Zhejiang Yunying Medical Technology Co., Ltd., Jiaxing, China
- KMID: 2547801
- DOI: http://doi.org/10.4143/crt.2023.415
Abstract
- Purpose
Loss-of-function mutations in the adenomatous polyposis coli (APC) gene are common in metastatic colorectal cancer (mCRC). However, the characteristic of APC specific mutations in mCRC is poorly understood. Here, we explored the clinical and molecular characteristics of N-terminal and C-terminal side APC mutations in Chinese patients with mCRC.
Materials and Methods
Hybrid capture-based next-generation sequencing was performed on tumor tissues from 275 mCRC pati-ents to detect mutations in 639 tumor-associated genes. The prognostic value and gene-pathway difference between APC specific mutations in mCRC patients were analyzed.
Results
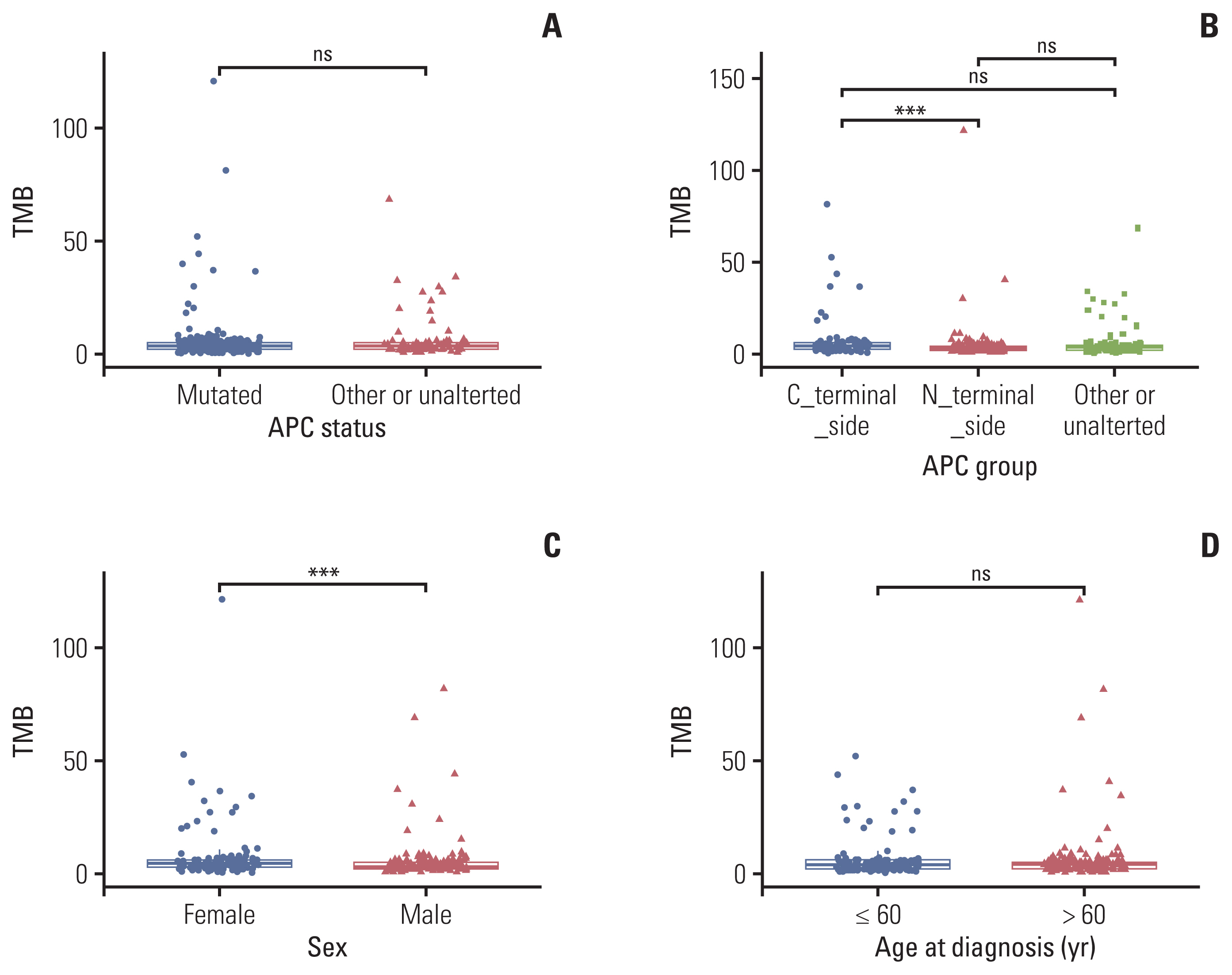
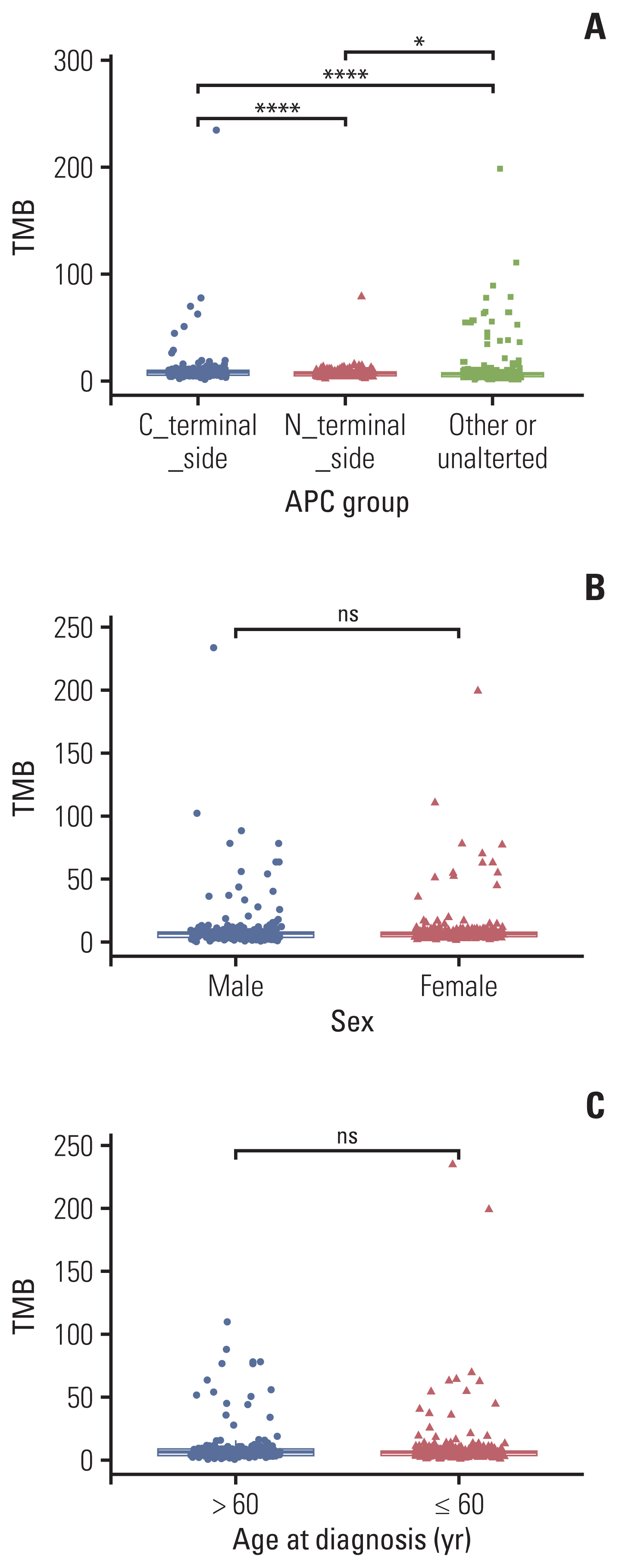
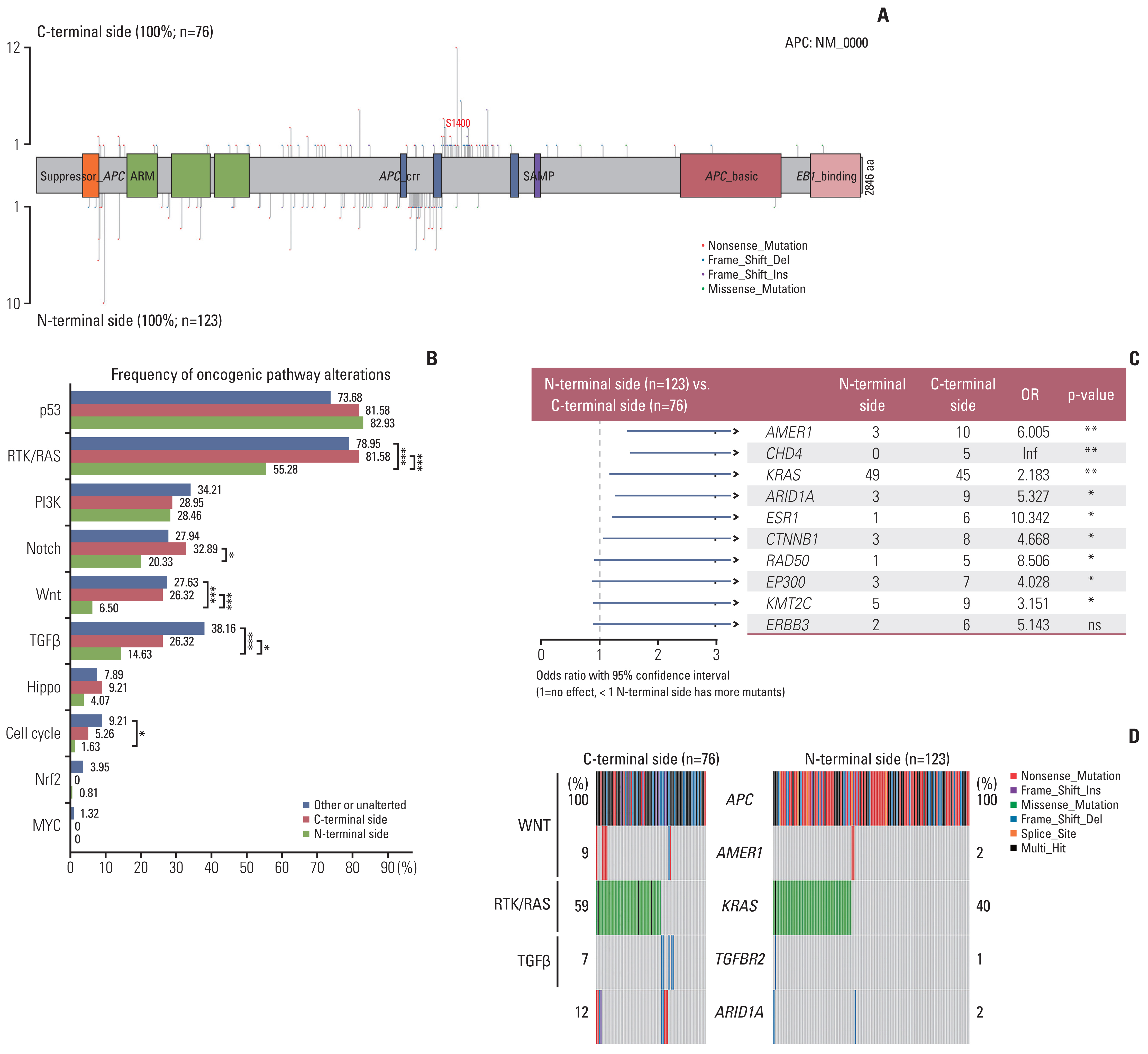
APC mutations were highly clustered, accounting for 73% of all mCRC patients, and most of them were truncating mutations. The tumor mutation burden of the N-terminal side APC mutations group (n=76) was significantly lower than that of the C-terminal side group (n=123) (p < 0.001), further confirmed by the public database. Survival analysis showed that mCRC patients with N-terminus side APC mutations had longer overall survival than C-terminus side. Tumor gene pathway analysis showed that gene mutations in the RTK/RAS, Wnt and transforming growth factor β signaling pathways of the C-terminal group were significantly higher than those of the N-terminal group (p < 0.05). Additionally, KRAS, AMER1, TGFBR2, and ARID1A driver mutations were more common in patients with C-terminal side APC mutations.
Conclusion
APC specific mutations have potential function as mCRC prognostic biomarkers. There are obvious differences in the gene mutation patterns between the C-terminus and N-terminus APC mutations group, which may have certain guiding significance for the subsequent precise treatment of mCRC.
Keyword
Figure
Reference
-
References
1. Bodmer WF, Bailey CJ, Bodmer J, Bussey HJ, Ellis A, Gorman P, et al. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature. 1987; 328:614–6.2. Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001; 1:55–67.3. Akiyama Y, Nagasaki H, Yagi KO, Nomizu T, Yuasa Y. Beta-catenin and adenomatous polyposis coli (APC) mutations in adenomas from hereditary non-polyposis colorectal cancer patients. Cancer Lett. 2000; 157:185–91.4. Lau T, Chan E, Callow M, Waaler J, Boggs J, Blake RA, et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013; 73:3132–44.5. Hankey W, Frankel WL, Groden J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting. Cancer Metastasis Rev. 2018; 37:159–72.6. Nelson SA, Li Z, Newton IP, Fraser D, Milne RE, Martin DM, et al. Tumorigenic fragments of APC cause dominant defects in directional cell migration in multiple model systems. Dis Model Mech. 2012; 5:940–7.7. Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. 2007; 120:3327–35.8. Mondaca S, Walch H, Nandakumar S, Chatila WK, Schultz N, Yaeger R. Specific mutations in APC, but not alterations in DNA damage response, associate with outcomes of patients with metastatic colorectal cancer. Gastroenterology. 2020; 159:1975–8.9. Cen B, Wei J, Wang D, Xiong Y, Shay JW, DuBois RN. Mutant APC promotes tumor immune evasion via PD-L1 in colorectal cancer. Oncogene. 2021; 40:5984–92.10. Lewis A, Davis H, Deheragoda M, Pollard P, Nye E, Jeffery R, et al. The C-terminus of Apc does not influence intestinal adenoma development or progression. J Pathol. 2012; 226:73–83.11. Han X, Zhang S, Zhou DC, Wang D, He X, Yuan D, et al. MSIsensor-ct: microsatellite instability detection using cfDNA sequencing data. Brief Bioinform. 2021; 22:bbaa402.12. Yamamoto D, Oshima H, Wang D, Takeda H, Kita K, Lei X, et al. Characterization of RNF43 frameshift mutations that drive Wnt ligand- and R-spondin-dependent colon cancer. J Pathol. 2022; 257:39–52.13. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.14. Lampropoulos P, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. TGF-beta signalling in colon carcinogenesis. Cancer Lett. 2012; 314:1–7.15. Staudacher JJ, Bauer J, Jana A, Tian J, Carroll T, Mancinelli G, et al. Activin signaling is an essential component of the TGF-β induced pro-metastatic phenotype in colorectal cancer. Sci Rep. 2017; 7:5569.16. Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2019; 29:44–65.17. Chiavarina B, Costanza B, Ronca R, Blomme A, Rezzola S, Chiodelli P, et al. Metastatic colorectal cancer cells maintain the TGFbeta program and use TGFBI to fuel angiogenesis. Theranostics. 2021; 11:1626–40.18. Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010; 28:466–74.19. Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol. 2017; 28:1023–31.20. Modest DP, Ricard I, Heinemann V, Hegewisch-Becker S, Schmiegel W, Porschen R, et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol. 2016; 27:1746–53.21. Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science. 2007; 316:1043–6.22. He H, Zhao X, Zhu Z, Du L, Chen E, Liu S, et al. MicroRNA-3191 promotes migration and invasion by downregulating TGFBR2 in colorectal cancer. J Biochem Mol Toxicol. 2019; 33:e22308.23. Zhang W, Zhang T, Jin R, Zhao H, Hu J, Feng B, et al. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J Exp Clin Cancer Res. 2014; 33:113.24. Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL, Zeng ZL, et al. Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer. World J Gastroenterol. 2014; 20:18404–12.25. Ye J, Zhou Y, Weiser MR, Gonen M, Zhang L, Samdani T, et al. Immunohistochemical detection of ARID1A in colorectal carcinoma: loss of staining is associated with sporadic microsatellite unstable tumors with medullary histology and high TNM stage. Hum Pathol. 2014; 45:2430–6.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Implications of Lymph Node Metastasis in Colorectal Cancer: Current Status and Future Perspectives
- Genetic Alterations and Microenvironment that Drive Malignant Progression of Colorectal Cancer: Lessons from Mouse and Organoid Models
- Understanding of molecular pathogenesis and genetic markers in colorectal cancer
- Paired Primary and Metastatic Tumor Analysis of Somatic Mutations in Synchronous and Metachronous Colorectal Cancer
- Longitudinal Comparative Analysis of Circulating Tumor DNA and Matched Tumor Tissue DNA in Patients with Metastatic Colorectal Cancer Receiving Palliative First-Line Systemic Anti-Cancer Therapy