Cancer Res Treat.
2024 Oct;56(4):1171-1182. 10.4143/crt.2024.016.
Longitudinal Comparative Analysis of Circulating Tumor DNA and Matched Tumor Tissue DNA in Patients with Metastatic Colorectal Cancer Receiving Palliative First-Line Systemic Anti-Cancer Therapy
- Affiliations
-
- 1Macrogen, Inc., Seoul, Korea
- 2Department of Internal Medicine, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- 3Biomedical Research Institute, Seoul National University Bundang Hospital, Seongnam, Korea
- 4Department of Statistics, Hankuk University of Foreign Studies, Yongin, Korea
- KMID: 2560251
- DOI: http://doi.org/10.4143/crt.2024.016
Abstract
- Purpose
This study aimed to compare tumor tissue DNA (ttDNA) and circulating tumor DNA (ctDNA) to explore the clinical applicability of ctDNA and to better understand clonal evolution in patients with metastatic colorectal cancer undergoing palliative first-line systemic therapy.
Materials and Methods
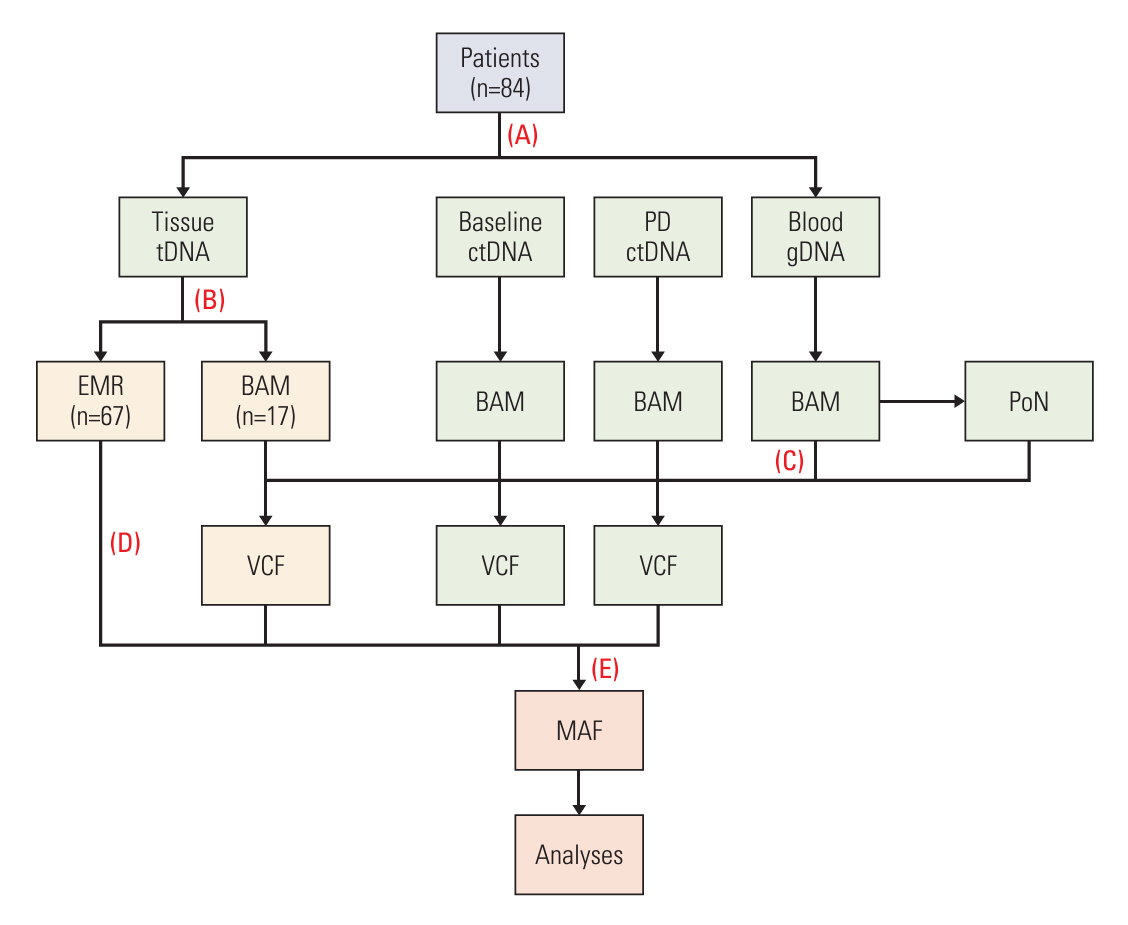
We performed targeted sequencing analysis of 88 cancer-associated genes using germline DNA, ctDNA at baseline (baseline-ctDNA), and ctDNA at progressive disease (PD-ctDNA). The results were compared with ttDNA data.
Results
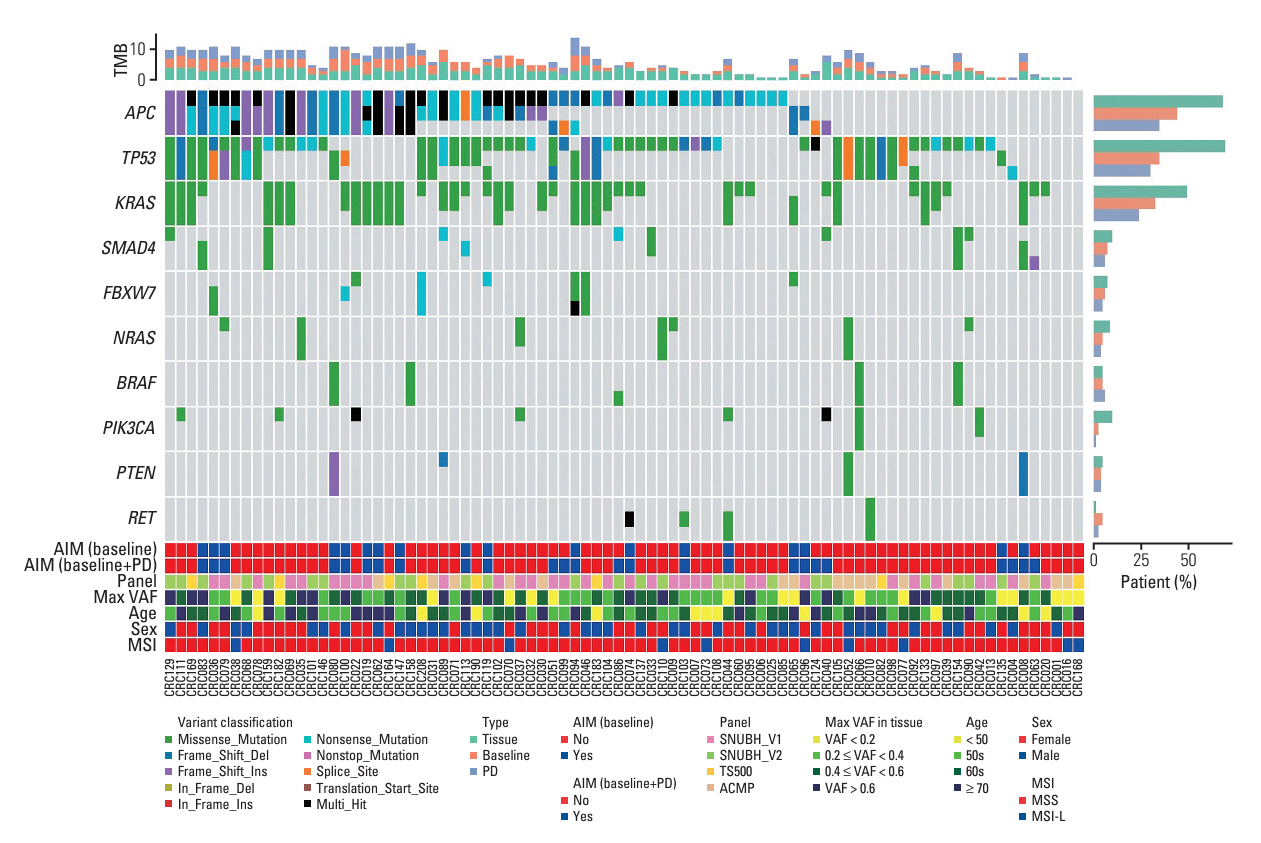
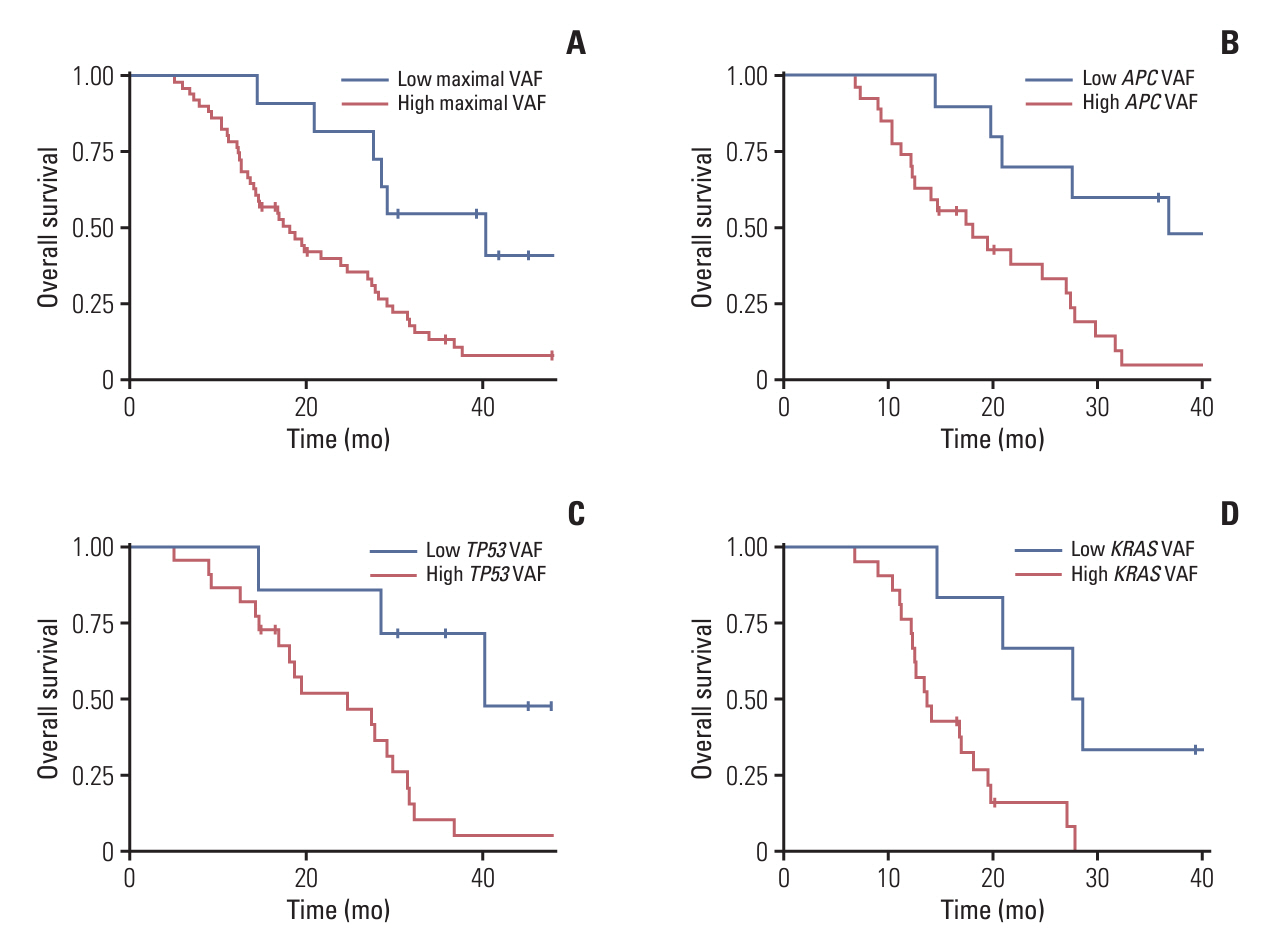
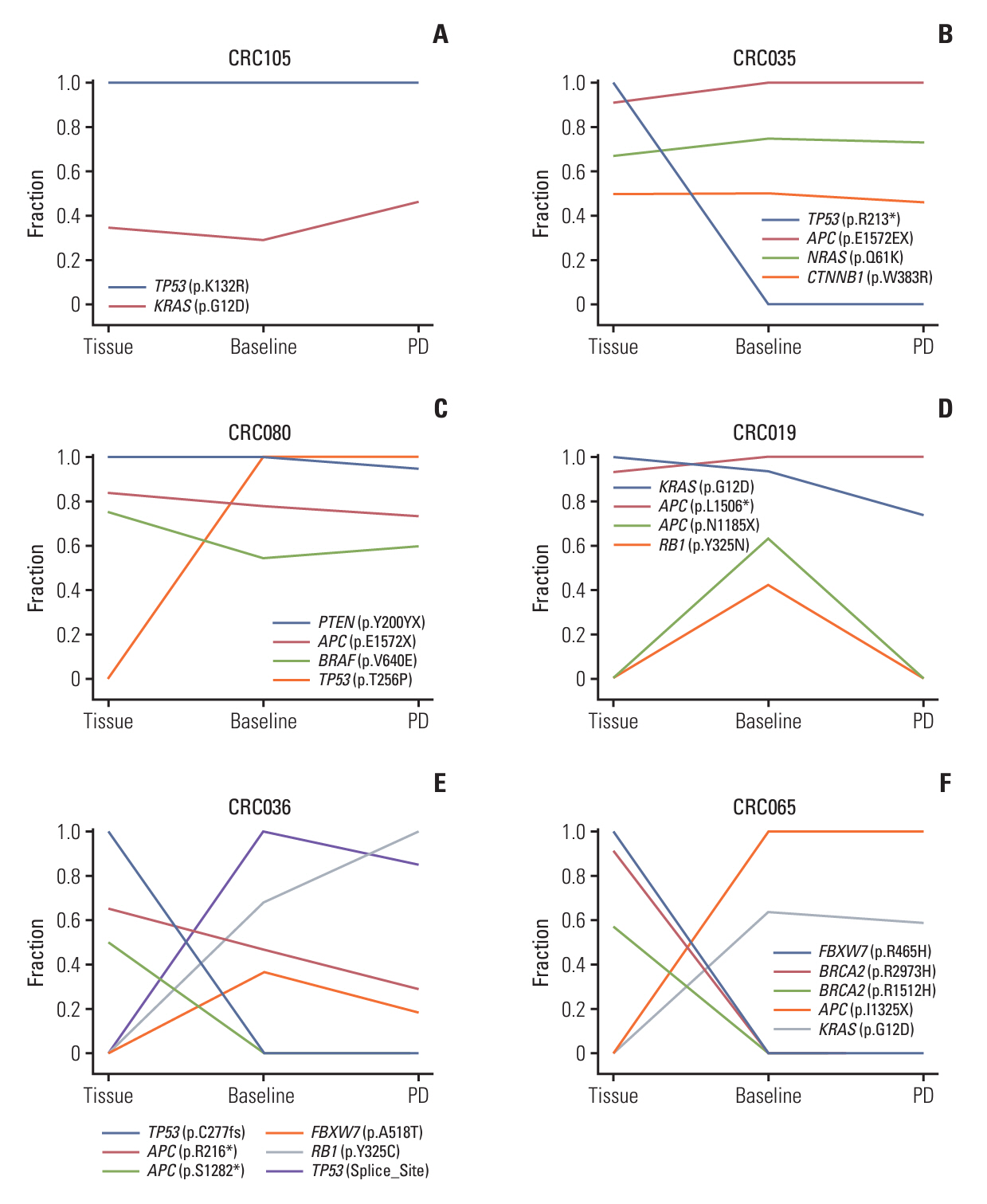
Among 208 consecutively enrolled patients, we selected 84 (41 males; median age, 59 years; range, 35 to 90 years) with all four sample types available. A total of 202 driver mutations were found in 34 genes. ttDNA exhibited the highest mutation frequency (n=232), followed by baseline-ctDNA (n=155) and PD-ctDNA (n=117). Sequencing ctDNA alongside ttDNA revealed additional mutations in 40 patients (47.6%). PD-ctDNA detected 13 novel mutations in 10 patients (11.9%) compared to ttDNA and baseline-ctDNA. Notably, seven mutations in five patients (6.0%) were missense or nonsense mutations in APC, TP53, SMAD4, and CDH1 genes. In baseline-ctDNA, higher maximal variant allele frequency (VAF) values (p=0.010) and higher VAF values of APC (p=0.012), TP53 (p=0.012), and KRAS (p=0.005) mutations were significantly associated with worse overall survival.
Conclusion
While ttDNA remains more sensitive than ctDNA, our ctDNA platform demonstrated validity and potential value when ttDNA was unavailable. Post-treatment analysis of PD-ctDNA unveiled new pathogenic mutations, signifying cancer’s clonal evolution. Additionally, baseline-ctDNA’s VAF values were prognostic after treatment.
Figure
Reference
-
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.
Article2. Kang MJ, Jung KW, Bang SH, Choi SH, Park EH, Yun EH, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2020. Cancer Res Treat. 2023; 55:385–99.
Article3. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.4. Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014; 15:1065–75.
Article5. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019; 381:1632–43.
Article6. Strickler JH, Cercek A, Siena S, Andre T, Ng K, Van Cutsem E, et al. Tucatinib plus trastuzumab for chemotherapy-refractory, HER2-positive, RAS wild-type unresectable or metastatic colorectal cancer (MOUNTAINEER): a multicentre, open-label, phase 2 study. Lancet Oncol. 2023; 24:496–508.7. Fakih MG, Salvatore L, Esaki T, Modest DP, Lopez-Bravo DP, Taieb J, et al. Sotorasib plus panitumumab in refractory colorectal cancer with mutated KRAS G12C. N Engl J Med. 2023; 389:2125–39.
Article8. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017; 168:613–28.
Article9. Turajlic S, Swanton C. Metastasis as an evolutionary process. Science. 2016; 352:169–75.
Article10. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013; 368:1199–209.
Article11. Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017; 17:223–38.
Article12. Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif. 2019; 17:100087.
Article13. Kim S, Cha Y, Lim Y, Roh H, Kang JK, Lee KH, et al. Mutational evolution after chemotherapy-progression in metastatic colorectal cancer revealed by circulating tumor DNA analysis. Int J Cancer. 2023; 153:571–83.14. Schwartz LH, Litiere S, de Vries E, Ford R, Gwyther S, Mandrekar S, et al. RECIST 1.1-Update and clarification: from the RECIST committee. Eur J Cancer. 2016; 62:132–7.
Article15. Hothorn T, Zeileis A. Generalized maximally selected statistics. Biometrics. 2008; 64:1263–9.
Article16. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–303.
Article17. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020; 581:434–43.
Article18. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016; 17:122.
Article19. Lee SB, Shin JY, Kwon NJ, Kim C, Seo JS. ClinPharmSeq: a targeted sequencing panel for clinical pharmacogenetics implementation. PLoS One. 2022; 17:e0272129.
Article20. Strickler JH, Loree JM, Ahronian LG, Parikh AR, Niedzwiecki D, Pereira AA, et al. Genomic landscape of cell-free DNA in patients with colorectal cancer. Cancer Discov. 2018; 8:164–73.
Article21. Boscolo Bielo L, Trapani D, Repetto M, Crimini E, Valenza C, Belli C, et al. Variant allele frequency: a decision-making tool in precision oncology? Trends Cancer. 2023; 9:1058–68.
Article22. Manca P, Corallo S, Lonardi S, Fuca G, Busico A, Leone AG, et al. Variant allele frequency in baseline circulating tumour DNA to measure tumour burden and to stratify outcomes in patients with RAS wild-type metastatic colorectal cancer: a translational objective of the Valentino study. Br J Cancer. 2022; 126:449–55.
Article23. Li M, Yang L, Hughes J, van den Hout A, Burns C, Woodhouse R, et al. Driver mutation variant allele frequency in circulating tumor DNA and association with clinical outcome in patients with non-small cell lung cancer and EGFR- and KRAS-mutated tumors. J Mol Diagn. 2022; 24:543–53.
Article24. Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014; 11:396–8.
Article25. Cox AD, Der CJ. Ras history: the saga continues. Small GTP ases. 2010; 1:2–27.26. De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010; 11:753–62.
Article27. McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015; 7:283ra54.
Article28. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015; 21:795–801.
Article29. Parseghian CM, Sun R, Woods M, Napolitano S, Lee HM, Alshenaifi J, et al. Resistance mechanisms to anti-epidermal growth factor receptor therapy in RAS/RAF wild-type colorectal cancer vary by regimen and line of therapy. J Clin Oncol. 2023; 41:460–71.30. Raghav K, Ou FS, Venook AP, Innocenti F, Sun R, Lenz HJ, et al. Acquired genomic alterations on first-line chemotherapy with cetuximab in advanced colorectal cancer: circulating tumor DNA analysis of the CALGB/SWOG-80405 trial (Alliance). J Clin Oncol. 2023; 41:472–8.
Article31. Topham JT, O’Callaghan CJ, Feilotter H, Kennecke HF, Lee YS, Li W, et al. Circulating tumor DNA identifies diverse landscape of acquired resistance to anti-epidermal growth factor receptor therapy in metastatic colorectal cancer. J Clin Oncol. 2023; 41:485–96.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Current Status of Chemotherapy in Colorectal Cancer: Updated Treatment Strategies
- Circulating Tumor Cell and Cell-free Circulating Tumor DNA in Lung Cancer
- Role of Liquid Biopsies in Colorectal Cancer
- Analysis of Circulating Tumor DNA to Predict Neoadjuvant Therapy Effectiveness and Breast Cancer Recurrence