Cancer Res Treat.
2017 Jan;49(1):161-167. 10.4143/crt.2015.490.
Paired Primary and Metastatic Tumor Analysis of Somatic Mutations in Synchronous and Metachronous Colorectal Cancer
- Affiliations
-
- 1Division of Oncology, Department of Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. twkimmd@amc.seoul.kr
- 2Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- 3Department of Colorectal Surgery, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2367513
- DOI: http://doi.org/10.4143/crt.2015.490
Abstract
- PURPOSE
Although the mutation status of KRAS is highly concordant in primary and metastatic lesions, it has not been generalized to other major pathway genes.
MATERIALS AND METHODS
In this study, 41 genes were evaluated and the mutational profiles were compared in 46 colorectal cancer patients with paired surgical specimens of primary and metastatic lesions: synchronous (n=27) and metachronous (n=19) lesions. A high-throughput mass spectrometry-based genotyping platform validated by orthogonal chemistry, OncoMap v.4.4, was used to evaluate the formalin-fixed, paraffin-embedded surgical specimens. The patients' demographics, tumor characteristics, and microsatellite instability status were analyzed by a retrospective chart review.
RESULTS
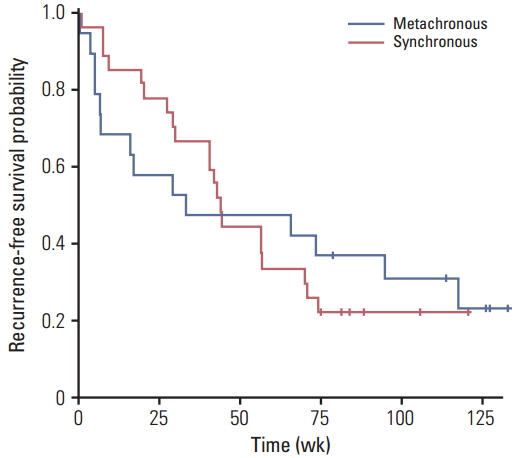
In this study,with OncoMap, mutationswere identified in 80.4% of patientswith the following frequency: KRAS (39.1%), TP53 (28.3%), APC (28.3%), PIK3CA (6.5%), BRAF (6.5%), and NRAS (4.3%). Although 19.6% (9/46) of the patients showed no gene mutations, 43.5% (20/46) and 37.0% (17/46) had mutations in one and two or more genes, respectively. The synchronous and metachronous lesions showed similar mutational profiles. Paired samples between primary and metastatic tumors differed in 7.4% (2/27) and 10.5% (2/19) for synchronous and metachronous according to OncoMap.
CONCLUSION
These findings indicate the major pathway genes, including KRAS, TP53, APC, PIK3CA, BRAF, and NRAS, are often concordant between the primary and metastatic lesions regardless of the temporal relationship of metastasis.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012; 366:883–92.
Article2. Mahmoud N, Bullard Dunn K. Metastasectomy for stage IV colorectal cancer. Dis Colon Rectum. 2010; 53:1080–92.
Article3. Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy? Genet Med. 2013; 15:517–27.4. Wright AA, Howitt BE, Myers AP, Dahlberg SE, Palescandolo E, Van Hummelen P, et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer. 2013; 119:3776–83.5. Kim YM, Lee SW, Chun SM, Kim DY, Kim JH, Kim KR, et al. Analysis and comparison of somatic mutations in paired primary and recurrent epithelial ovarian cancer samples. PLoS One. 2014; 9:e99451.
Article6. Mao C, Wu XY, Yang ZY, Threapleton DE, Yuan JQ, Yu YY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep. 2015; 5:8065.
Article7. Knijn N, Mekenkamp LJ, Klomp M, Vink-Borger ME, Tol J, Teerenstra S, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011; 104:1020–6.
Article8. Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012; 486:532–6.
Article9. Kawamoto Y, Tsuchihara K, Yoshino T, Ogasawara N, Kojima M, Takahashi M, et al. KRAS mutations in primary tumours and post-FOLFOX metastatic lesions in cases of colorectal cancer. Br J Cancer. 2012; 107:340–4.
Article10. Lee SY, Haq F, Kim D, Jun C, Jo HJ, Ahn SM, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One. 2014; 9:e90459.
Article11. Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014; 15:454.
Article12. Ghiringhelli F, Hennequin A, Drouillard A, Lepage C, Faivre J, Bouvier AM. Epidemiology and prognosis of synchronous and metachronous colon cancer metastases: a French population-based study. Dig Liver Dis. 2014; 46:854–8.
Article13. Chu J, Goktepe O, Cheung WY. Natural history and outcomes of synchronous and metachronous colorectal cancers (CRC): a population-based analysis. J Clin Oncol. 2013; 31(Suppl):Abstr 3593.
Article14. Slesser AA, Georgiou P, Brown G, Mudan S, Goldin R, Tekkis P. The tumour biology of synchronous and metachronous colorectal liver metastases: a systematic review. Clin Exp Metastasis. 2013; 30:457–70.
Article15. Rose JS, Serna DS, Martin LK, Li X, Weatherby LM, AbdelMisih S, et al. Influence of KRAS mutation status in metachronous and synchronous metastatic colorectal adenocarcinoma. Cancer. 2012; 118:6243–52.16. El Zouhairi M, Charabaty A, Pishvaian MJ. Molecularly targeted therapy for metastatic colon cancer: proven treatments and promising new agents. Gastrointest Cancer Res. 2011; 4:15–21.17. Jiang JK, Chen YJ, Lin CH, Yu IT, Lin JK. Genetic changes and clonality relationship between primary colorectal cancers and their pulmonary metastases: an analysis by comparative genomic hybridization. Genes Chromosomes Cancer. 2005; 43:25–36.18. Miranda E, Bianchi P, Destro A, Morenghi E, Malesci A, Santoro A, et al. Genetic and epigenetic alterations in primary colorectal cancers and related lymph node and liver metastases. Cancer. 2013; 119:266–76.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Oncologic outcomes following metastasectomy in colorectal cancer patients developing distant metastases after initial treatment
- The Clinicopathologic Features of Synchronous and Metachronous Cancer in Patients with Gastric Cancer
- Treatment of Multiple Colorectal Cancers
- Clinical Characteristics of Synchronous Adenomas Designated as Risk Factors of Metachronous Malignancy in Colorectal Cancer
- Clinicopathologic characteristics and survival rate in patients with synchronous or metachronous double primary colorectal and gastric cancer