Ann Lab Med.
2020 Jan;40(1):15-20. 10.3343/alm.2020.40.1.15.
Development of Tigecycline Resistance in Carbapenemase-Producing Klebsiella pneumoniae Sequence Type 147 via AcrAB Overproduction Mediated by Replacement of the ramA Promoter
- Affiliations
-
- 1Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, Korea. kscpjsh@yuhs.ac
- KMID: 2457488
- DOI: http://doi.org/10.3343/alm.2020.40.1.15
Abstract
- BACKGROUND
Carbapenem-resistant K. pneumoniae 2297, isolated from a patient treated with tigecycline for pneumonia, developed tigecycline resistance, in contrast to carbapenem-resistant isolate 1215, which was collected four months prior to the 2297 isolate. Mechanisms underlying tigecycline resistance were elucidated for the clinical isolates.
METHODS
The tigecycline minimum inhibitory concentration (MIC) was determined using the broth microdilution method, with or without phenylalanine-arginine β-naphthylamide (PABN), and whole-genome sequencing was carried out by single-molecule real-time sequencing. The expression levels of the genes acrA, oqxA, ramA, rarA, and rpoB were determined by reverse-transcription quantitative PCR.
RESULTS
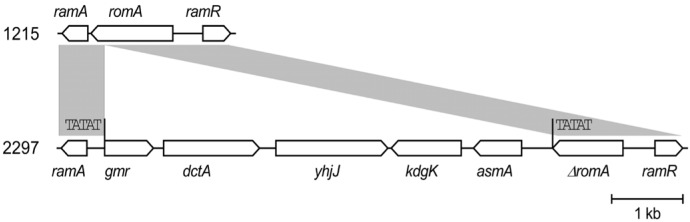
Both isolates presented identical antibiograms, except for tigecycline, which showed an MIC of 0.5 mg/L in 1215 and 2 mg/L in 2297. The addition of PABN to tigecycline-resistant 2297 caused a four-fold decrease in the tigecycline MIC to 0.5 mg/L, although acrA expression (encoding the AcrAB efflux pump) was upregulated by 2.5 fold and ramA expression (encoding the pump activator RamA) was upregulated by 1.4 fold. We identified a 6,096-bp fragment insertion flanking direct TATAT repeats that disrupted the romA gene located upstream of ramA in the chromosome of K. pneumoniae 2297; the insertion led the ramA gene promoter replacement resulting in stronger activation of the gene.
CONCLUSIONS
The K. pneumoniae isolate developed tigecycline resistance during tigecycline treatment. It was related to the overexpression of the AcrAB resistance-nodulation-cell division efflux system due to promoter replacement.
Keyword
MeSH Terms
Figure
Reference
-
1. Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013; 11:297–298. PMID: 23458769.2. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015; 59:5873–5884. PMID: 26169401.3. Livermore DM. Tigecycline: what is it, and where should it be used? J Antimicrob Chemother. 2005; 56:611–614. PMID: 16120626.4. Dean CR, Visalli MA, Projan SJ, Sum PE, Bradford PA. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother. 2003; 47:972–978. PMID: 12604529.5. Hornsey M, Ellington MJ, Doumith M, Hudson S, Livermore DM, Woodford N. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J Antimicrob Chemother. 2010; 65:479–482. PMID: 20051474.6. Veleba M, De Majumdar S, Hornsey M, Woodford N, Schneiders T. Genetic characterization of tigecycline resistance in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Antimicrob Chemother. 2013; 68:1011–1018. PMID: 23349441.7. Hirata T, Saito A, Nishino K, Tamura N, Yamaguchi A. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob Agents Chemother. 2004; 48:2179–2184. PMID: 15155219.8. Veleba M, Schneiders T. Tigecycline resistance can occur independently of the ramA gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2012; 56:4466–4467. PMID: 22644034.9. Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2005; 49:1017–1022. PMID: 15728897.10. De Majumdar S, Veleba M, Finn S, Fanning S, Schneiders T. Elucidating the regulon of multidrug resistance regulator RarA in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013; 57:1603–1609. PMID: 23318802.11. Cho SY, Huh HJ, Baek JY, Chung NY, Ryu JG, Ki CS, et al. Klebsiella pneumoniae co-producing NDM-5 and OXA-181 carbapenemases, South Korea. Emerg Infect Dis. 2015; 21:1088–1089. PMID: 25988911.12. Du X, He F, Shi Q, Zhao F, Xu J, Fu Y, et al. The Rapid Emergence of Tigecycline Resistance in blaKPC-2 Harboring Klebsiella pneumoniae, as Mediated in Vivo by Mutation in tetA During Tigecycline Treatment. Front Microbiol. 2018; 9:648. PMID: 29675006.13. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43:4178–4182. PMID: 16081970.14. EUCAST. Antimicrobial susceptibility testing of bacteria. Updated on Jan 2017. http://www.eucast.org/ast_of_bacteria/.15. EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. version 7.1. Updated on Mar 2017. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.16. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011; 70:119–123. PMID: 21398074.17. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. PMID: 9254694.18. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012; 67:2640–2644. PMID: 22782487.19. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014; 58:3895–3903. PMID: 24777092.20. Komatsu T, Ohta M, Kido N, Arakawa Y, Ito H, Kato N. Increased resistance to multiple drugs by introduction of the Enterobacter cloacae romA gene into OmpF porin-deficient mutants of Escherichia coli K-12. Antimicrob Agents Chemother. 1991; 35:2155–2158. PMID: 1662028.21. Rosenblum R, Khan E, Gonzalez G, Hasan R, Schneiders T. Genetic regulation of the ramA locus and its expression in clinical isolates of Klebsiella pneumoniae. Int J Antimicrob Agents. 2011; 38:39–45. PMID: 21514798.22. Bauer G, Berens C, Projan SJ, Hillen W. Comparison of tetracycline and tigecycline binding to ribosomes mapped by dimethylsulphate and drug-directed Fe2+ cleavage of 16S rRNA. J Antimicrob Chemother. 2004; 53:592–599. PMID: 14985271.23. Fang L, Chen Q, Shi K, Li X, Shi Q, He F, et al. Step-Wise increase in tigecycline resistance in Klebsiella pneumoniae associated with mutations in ramR, lon and rpsJ. PLoS One. 2016; 11:e0165019. PMID: 27764207.24. van Duin D, Cober ED, Richter SS, Perez F, Cline M, Kaye KS, et al. Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin Microbiol Infect. 2014; 20:O1117–O1120. PMID: 24931918.25. Nigo M, Cevallos CS, Woods K, Flores VM, Francis G, Perlman DC, et al. Nested case-control study of the emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013; 57:5743–5746. PMID: 23979745.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Isolation of a Klebsiella pneumoniae Isolate of Sequence Type 258 Producing KPC-2 Carbapenemase in Korea
- The Resistance Mechanism and Clonal Distribution of Tigecycline-Nonsusceptible Klebsiella pneumoniae Isolates in Korea
- Antimicrobial Resistance Caused by KPC-2 Encoded by Promiscuous Plasmids of the Klebsiella pneumoniae ST307 Strain
- Two Cases of Neonatal Osteomyelitis due to Extended Spectrum beta-lactamase Producing Klebsiella pneumoniae
- Prevalence and Molecular Characteristics of Carbapenemase-Producing Enterobacteriaceae From Five Hospitals in Korea