Diabetes Metab J.
2019 Aug;43(4):422-431. 10.4093/dmj.2018.0090.
Additional Effect of Dietary Fiber in Patients with Type 2 Diabetes Mellitus Using Metformin and Sulfonylurea: An Open-Label, Pilot Trial
- Affiliations
-
- 1Division of Endocrinology and Metabolism, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. leemk@skku.edu
- 2Department of Environmental Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea.
- 3Department of Endocrinology and Metabolism, Kyung Hee University Hospital at Gangdong, Kyung Hee University School of Medicine, Seoul, Korea.
- 4Division of Endocrinology and Metabolism, Department of Medicine, Korea University Guro Hospital, Korea University College of Medicine, Seoul, Korea.
- KMID: 2455666
- DOI: http://doi.org/10.4093/dmj.2018.0090
Abstract
- BACKGROUND
Metformin, sulfonylurea, and dietary fiber are known to affect gut microbiota in patients with type 2 diabetes mellitus (T2DM). This open and single-arm pilot trial investigated the effects of the additional use of fiber on glycemic parameters, insulin, incretins, and microbiota in patients with T2DM who had been treated with metformin and sulfonylurea.
METHODS
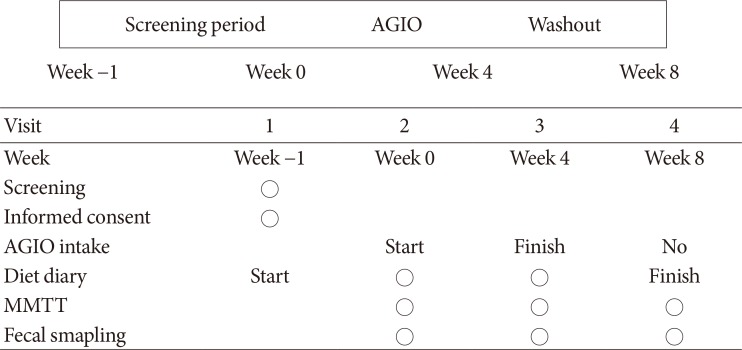
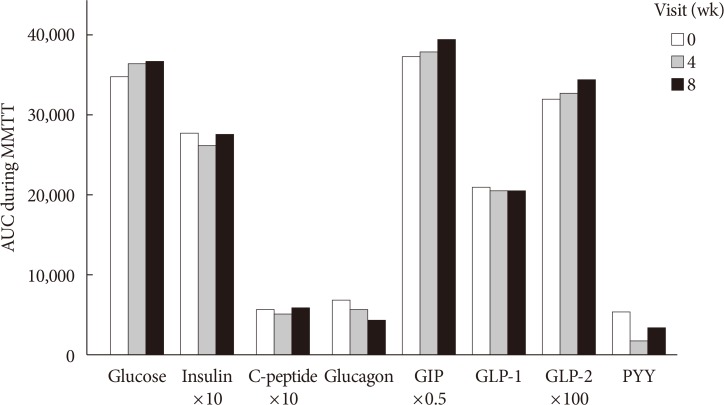
Participants took fiber for 4 weeks and stopped for the next 4 weeks. Glycemic parameters, insulin, incretins during mixed-meal tolerance test (MMTT), lipopolysaccharide (LPS) level, and fecal microbiota were analyzed at weeks 0, 4, and 8. The first tertile of difference in glucose area under the curve during MMTT between weeks 0 and 4 was defined as "˜responders' and the third as "˜nonresponders,' respectively.
RESULTS
In all 10 participants, the peak incretin levels during MMTT were higher and LPS were lower at week 4 as compared with at baseline. While the insulin sensitivity of the "˜responders' increased at week 4, that of the "˜nonresponders' showed opposite results. However, the results were not statistically significant. In all participants, metabolically unfavorable microbiota decreased at week 4 and were restored at week 8. At baseline, metabolically hostile bacteria were more abundant in the "˜nonresponders.' In "˜responders,' Roseburia intestinalis increased at week 4.
CONCLUSION
While dietary fiber did not induce additional changes in glycemic parameters, it showed a trend of improvement in insulin sensitivity in "˜responders.' Even if patients are already receiving diabetes treatment, the additional administration of fiber can lead to additional benefits in the treatment of diabetes.
MeSH Terms
Figure
Reference
-
1. American Diabetes Association. Standards of medical care in diabetes: 2018. Diabetes Care. 2018; 41(Suppl 1):S1–S153. PMID: 29222369.2. Abutair AS, Naser IA, Hamed AT. Soluble fibers from psyllium improve glycemic response and body weight among diabetes type 2 patients (randomized control trial). Nutr J. 2016; 15:86. PMID: 27733151.
Article3. Bajorek SA, Morello CM. Effects of dietary fiber and low glycemic index diet on glucose control in subjects with type 2 diabetes mellitus. Ann Pharmacother. 2010; 44:1786–1792. PMID: 20959501.
Article4. Gibb RD, McRorie JW Jr, Russell DA, Hasselblad V, D'Alessio DA. Psyllium fiber improves glycemic control proportional to loss of glycemic control: a meta-analysis of data in euglycemic subjects, patients at risk of type 2 diabetes mellitus, and patients being treated for type 2 diabetes mellitus. Am J Clin Nutr. 2015; 102:1604–1614. PMID: 26561625.
Article5. Hall M, Flinkman T. Do fiber and psyllium fiber improve diabetic metabolism? Consult Pharm. 2012; 27:513–516. PMID: 22910133.
Article6. Pastors JG, Blaisdell PW, Balm TK, Asplin CM, Pohl SL. Psyllium fiber reduces rise in postprandial glucose and insulin concentrations in patients with non-insulin-dependent diabetes. Am J Clin Nutr. 1991; 53:1431–1435. PMID: 1852093.
Article7. Weickert MO, Mohlig M, Koebnick C, Holst JJ, Namsolleck P, Ristow M, Osterhoff M, Rochlitz H, Rudovich N, Spranger J, Pfeiffer AF. Impact of cereal fibre on glucose-regulating factors. Diabetologia. 2005; 48:2343–2353. PMID: 16172868.
Article8. Slavin JL. Position of the American Dietetic Association: health implications of dietary fiber. J Am Diet Assoc. 2008; 108:1716–1731. PMID: 18953766.9. Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2014; 5:202–207. PMID: 24429443.
Article10. Esteve E, Ricart W, Fernandez-Real JM. Gut microbiota interactions with obesity, insulin resistance and type 2 diabetes: did gut microbiote co-evolve with insulin resistance? Curr Opin Clin Nutr Metab Care. 2011; 14:483–490. PMID: 21681087.11. Han JL, Lin HL. Intestinal microbiota and type 2 diabetes: from mechanism insights to therapeutic perspective. World J Gastroenterol. 2014; 20:17737–17745. PMID: 25548472.
Article12. Hur KY, Lee MS. Gut microbiota and metabolic disorders. Diabetes Metab J. 2015; 39:198–203. PMID: 26124989.
Article13. Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015; 7:2839–2849. PMID: 25875123.
Article14. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jorgensen T, Levenez F, Dore J; MetaHIT consortium, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015; 528:262–266. PMID: 26633628.
Article15. Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol. 2014; 80:5935–5943. PMID: 25038099.
Article16. Mardinoglu A, Boren J, Smith U. Confounding effects of metformin on the human gut microbiome in type 2 diabetes. Cell Metab. 2016; 23:10–12. PMID: 26771114.
Article17. Napolitano A, Miller S, Nicholls AW, Baker D, Van Horn S, Thomas E, Rajpal D, Spivak A, Brown JR, Nunez DJ. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One. 2014; 9:e100778. PMID: 24988476.
Article18. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, Stahlman M, Olsson LM, Serino M, Planas-Felix M, Xifra G, Mercader JM, Torrents D, Burcelin R, Ricart W, Perkins R, Fernandez-Real JM, Backhed F. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017; 23:850–858. PMID: 28530702.
Article19. Huo T, Xiong Z, Lu X, Cai S. Metabonomic study of biochemical changes in urinary of type 2 diabetes mellitus patients after the treatment of sulfonylurea antidiabetic drugs based on ultra-performance liquid chromatography/mass spectrometry. Biomed Chromatogr. 2015; 29:115–122. PMID: 24890121.
Article20. Montandon SA, Jornayvaz FR. Effects of antidiabetic drugs on gut microbiota composition. Genes (Basel). 2017; 8.
Article21. Korean Diabetes Association. Diabetes Fact Sheet in Korea 2018. Seoul: Korean Diabetes Association;2018.22. Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999; 22:1462–1470. PMID: 10480510.
Article23. Levy JC, Matthews DR, Hermans MP. Correct homeostasis model assessment (HOMA) evaluation uses the computer program. Diabetes Care. 1998; 21:2191–2192. PMID: 9839117.
Article24. Katz A, Nambi SS, Mather K, Baron AD, Follmann DA, Sullivan G, Quon MJ. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab. 2000; 85:2402–2410. PMID: 10902785.
Article25. Hur M, Kim Y, Song HR, Kim JM, Choi YI, Yi H. Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microbiol. 2011; 77:7611–7619. PMID: 21890678.
Article26. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011; 27:2194–2200. PMID: 21700674.
Article27. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62(Pt 3):716–721. PMID: 22140171.
Article28. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011; 12:R60. PMID: 21702898.
Article29. Puddu A, Sanguineti R, Montecucco F, Viviani GL. Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediators Inflamm. 2014; 2014:162021. PMID: 25214711.
Article30. Woting A, Blaut M. The intestinal microbiota in metabolic disease. Nutrients. 2016; 8:202. PMID: 27058556.
Article31. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003; 278:11312–11319. PMID: 12496283.
Article32. Mattace Raso G, Simeoli R, Russo R, Iacono A, Santoro A, Paciello O, Ferrante MC, Canani RB, Calignano A, Meli R. Effects of sodium butyrate and its synthetic amide derivative on liver inflammation and glucose tolerance in an animal model of steatosis induced by high fat diet. PLoS One. 2013; 8:e68626. PMID: 23861927.
Article33. Roelofsen H, Priebe MG, Vonk RJ. The interaction of short-chain fatty acids with adipose tissue: relevance for prevention of type 2 diabetes. Benef Microbes. 2010; 1:433–437. PMID: 21831781.
Article34. Romeo GR, Lee J, Shoelson SE. Metabolic syndrome, insulin resistance, and roles of inflammation: mechanisms and therapeutic targets. Arterioscler Thromb Vasc Biol. 2012; 32:1771–1776. PMID: 22815343.35. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490:55–60. PMID: 23023125.
Article36. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012; 143:913–916. PMID: 22728514.
Article37. Neyrinck AM, Possemiers S, Verstraete W, De Backer F, Cani PD, Delzenne NM. Dietary modulation of clostridial cluster XIVa gut bacteria (Roseburia spp.) by chitin-glucan fiber improves host metabolic alterations induced by high-fat diet in mice. J Nutr Biochem. 2012; 23:51–59. PMID: 21411304.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Letter: Efficacy and Safety of Voglibose Plus Metformin in Patients with Type 2 Diabetes Mellitus: A Randomized Controlled Trial (Diabetes Metab J 2019;43;276-86)
- Combination Therapy of Oral Hypoglycemic Agents in Patients with Type 2 Diabetes Mellitus
- The Effect of Rosiglitazone and Metformin Therapy, as an Initial Therapy, in Patients with Type 2 Diabetes Mellitus
- The Effect of Metformin in Obese Pediatric Patients with Type 2 Diabetes
- Comparison of Antidiabetic Regimens in Patients with Type 2 Diabetes Uncontrolled by Combination Therapy of Sulfonylurea and Metformin: Results of the MOHAS Disease Registry in Korea