Ann Surg Treat Res.
2019 Apr;96(4):153-161. 10.4174/astr.2019.96.4.153.
Genetic features associated with ¹â¸F-FDG uptake in intrahepatic cholangiocarcinoma
- Affiliations
-
- 1Department of Surgery, Keimyung University Dongsan Medical Center, Keimyung University School of Medicine, Daegu, Korea. ahnksmd@gmail.com
- 2Institute for Cancer Research, Keimyung University, Daegu, Korea.
- 3Department of Nuclear Medicine, Keimyung University Dongsan Medical Center, Daegu, Korea.
- 4Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN, USA.
- 5Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN, USA.
- KMID: 2442209
- DOI: http://doi.org/10.4174/astr.2019.96.4.153
Abstract
- PURPOSE
In intrahepatic cholangiocarcinoma (iCCA), genetic characteristics on ¹â¸F-fluorodeoxyglucose (¹â¸F-FDG)-PET scans are not yet clarified. If specific genetic characteristics were found to be related to FDG uptake in iCCA, we can predict molecular features based on the FDG uptake patterns and to distinguish different types of treatments. In this purpose, we analyzed RNA sequencing in iCCA patients to evaluate gene expression signatures associated with FDG uptake patterns.
METHODS
We performed RNA sequencing of 22 cases iCCA who underwent preoperative ¹â¸F-FDG-PET, and analyzed the clinical and molecular features according to the maximum standard uptake value (SUVmax). Genes and biological pathway which are associated with SUVmax were analyzed.
RESULTS
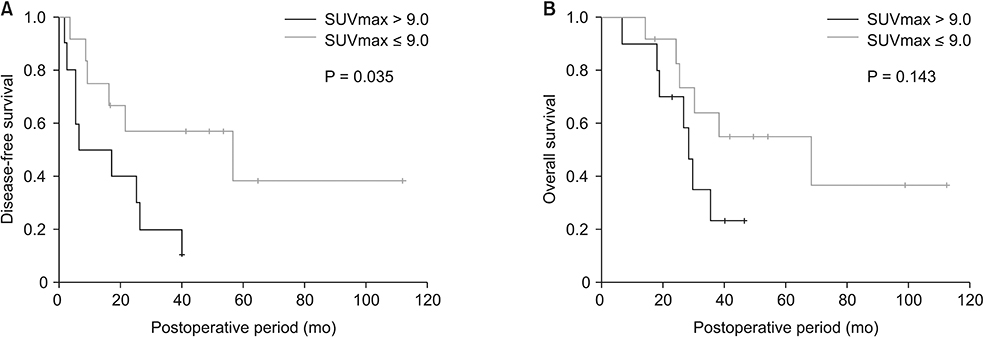
Patients with SUVmax higher than 9.0 (n = 9) had poorer disease-free survival than those with lower SUVmax (n = 13, P = 0.035). Genes related to glycolysis and gluconeogenesis, phosphorylation and cell cycle were significantly correlated with SUVmax (r ≥ 0.5). RRM2, which is related to the toxicity of Gemcitabine was positively correlated with SUVmax, and SLC27A2 which is associated with Cisplastin response was negatively correlated with SUVmax. According to the pathway analysis, cell cycle, cell division, hypoxia, inflammatory, and metabolism-related pathways were enriched in high SUVmax patients.
CONCLUSION
The genomic features of gene expression and pathways can be predicted by FDG uptake features in iCCA. Patients with high FDG uptake have enriched cell cycle, metabolism and hypoxic pathways, which may lead to a more rational targeted treatment approach.
Keyword
MeSH Terms
Figure
Reference
-
1. Patel T. Worldwide trends in mortality from biliary tract malignancies. BMC Cancer. 2002; 2:10.
Article2. Hu JH, Tang JH, Lin CH, Chu YY, Liu NJ. Preoperative staging of cholangiocarcinoma and biliary carcinoma using 18F-fluorodeoxyglucose positron emission tomography: a meta-analysis. J Investig Med. 2018; 66:52–61.
Article3. Jiang L, Tan H, Panje CM, Yu H, Xiu Y, Shi H. Role of 18F-FDG PET/CT imaging in intrahepatic cholangiocarcinoma. Clin Nucl Med. 2016; 41:1–7.
Article4. Lee Y, Yoo IR, Boo SH, Kim H, Park HL, Hyun OJ. The role of F-18 FDG PET/CT in intrahepatic cholangiocarcinoma. Nucl Med Mol Imaging. 2017; 51:69–78.
Article5. Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012; 142:1021–1031.e15.
Article6. Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013; 144:829–840.
Article7. Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017; 7:1116–1135.
Article8. Ito T, Sakurai-Yageta M, Goto A, Pairojkul C, Yongvanit P, Murakami Y. Genomic and transcriptional alterations of cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014; 21:380–387.
Article9. Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. J Thorac Oncol. 2008; 3:6–12.
Article10. Heiden BT, Chen G, Hermann M, Brown RKJ, Orringer MB, Lin J, et al. 18F-FDG PET intensity correlates with a hypoxic gene signature and other oncogenic abnormalities in operable non-small cell lung cancer. PLoS One. 2018; 13:e0199970.
Article11. Ahn SG, Lee JH, Lee HW, Jeon TJ, Ryu YH, Kim KM, et al. Comparison of standardized uptake value of 18F-FDG-PET-CT with 21-gene recurrence score in estrogen receptor-positive, HER2-negative breast cancer. PLoS One. 2017; 12:e0175048.
Article12. Ikeno Y, Seo S, Iwaisako K, Yoh T, Nakamoto Y, Fuji H, et al. Preoperative metabolic tumor volume of intrahepatic cholangiocarcinoma measured by 18F-FDG-PET is associated with the KRAS mutation status and prognosis. J Transl Med. 2018; 16:95.
Article13. Kwee SA, Okimoto GS, Chan OT, Tiirikainen M, Wong LL. Metabolic characteristics distinguishing intrahepatic cholangiocarcinoma: a negative pilot study of (18)F-fluorocholine PET/CT clarified by transcriptomic analysis. Am J Nucl Med Mol Imaging. 2016; 6:73–83.14. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010; 17:1471–1474.
Article15. Kalari KR, Nair AA, Bhavsar JD, O'Brien DR, Davila JI, Bockol MA, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 2014; 15:224.
Article16. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36.
Article17. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25.
Article18. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930.
Article19. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550.
Article20. Alvarellos ML, Lamba J, Sangkuhl K, Thorn CF, Wang L, Klein DJ, et al. PharmGKB summary: gemcitabine pathway. Pharmacogenet Genomics. 2014; 24:564–574.21. Su J, Wu S, Tang W, Qian H, Zhou H, Guo T. Reduced SLC27A2 induces cisplatin resistance in lung cancer stem cells by negatively regulating Bmi1-ABCG2 signaling. Mol Carcinog. 2016; 55:1822–1832.
Article22. Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 2017; 18:2780–2794.23. Kaplon J, van Dam L, Peeper D. Two-way communication between the metabolic and cell cycle machineries: the molecular basis. Cell Cycle. 2015; 14:2022–2032.
Article24. Challapalli A, Aboagye EO. Positron emission tomography imaging of tumor cell metabolism and application to therapy response monitoring. Front Oncol. 2016; 6:44.
Article25. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008; 7:11–20.
Article26. Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008; 105:18782–18787.
Article27. Ke F, Wang Z, Song X, Ma Q, Hu Y, Jiang L, et al. Cryptotanshinone induces cell cycle arrest and apoptosis through the JAK2/STAT3 and PI3K/Akt/NFκB pathways in cholangiocarcinoma cells. Drug Des Devel Ther. 2017; 11:1753–1766.28. Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011; 71:6921–6925.29. Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem. 2002; 277:23111–23115.
Article30. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009; 324:1029–1033.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Role of ¹â¸F-FDG PET-CT in Monitoring the Cyclophosphamide Induced Pulmonary Toxicity in Patients with Breast Cancer: 2 Case Reports
- Different 18F-FDG Uptake According to Tumor Location and Morphology of Cholangiocarcinoma and Its Clinical Implication
- 68Ga-FAPI-46 PET/MR Detects Recurrent Cholangiocarcinoma and Intraductal Papillary Mucinous Neoplasm in a Patient Showing Increasing CEA with Negative 18F-FDG PET/CT and Conventional CT
- The role of ¹â¸F-fluorodeoxyglucose positron emission tomography in the assessment of disease activity of adult-onset Still’s disease
- Recent Updates in the Imaging Diagnosis of Cholangiocarcinoma