Ann Clin Microbiol.
2019 Mar;22(1):1-8. 10.5145/ACM.2019.22.1.1.
Differences in Antimicrobial Resistance Phenotypes by the Group of CTX-M Extended-Spectrum β-Lactamase
- Affiliations
-
- 1Department of Clinical Pathology, Sangji University College of Science, Wonju, Korea.
- 2Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, Korea. ejyoon@yuhs.ac
- 3Department of Laboratory Medicine, Chonnam National University School of Medicine, Gwangju, Korea.
- 4Department of Laboratory Medicine, Inje University Busan Paik Hospital, Busan, Korea.
- 5Department of Laboratory Medicine, College of Medicine, Chungbuk National University, Cheongju, Korea.
- 6Department of Laboratory Medicine, National Health Insurance Service Ilsan Hospital, Goyang, Korea.
- 7Department of Laboratory Medicine, Wonju Severance Christian Hospital, Yonsei University Wonju College of Medicine, Wonju, Korea.
- 8Department of Laboratory Medicine, Hallym University College of Medicine, Hwaseong, Korea.
- 9Department of Laboratory Medicine, School of Medicine, Jeju National University, Jeju, Korea.
- KMID: 2441650
- DOI: http://doi.org/10.5145/ACM.2019.22.1.1
Abstract
- BACKGROUND
Escherichia coli and Klebsiella pneumoniae clinical isolates producing CTX-M extendedspectrum β-lactamases (ESBLs) were assessed for antimicrobial resistance phenotypes varied by group of enzymes.
METHODS
A total of 1,338 blood isolates, including 959 E. coli and 379 K. pneumoniae, were studied. All the strains were collected between January and July 2017 from eight general hospitals in South Korea. The species were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Antimicrobial susceptibilities were determined by disk diffusion methods and ESBL phenotypes by double-disk synergy tests using disks containing cefotaxime, ceftazidime, cefepime, aztreonam, and clavulanic acid (CA). The genes for β-lactamases were identified by PCR and sequencing.
RESULTS
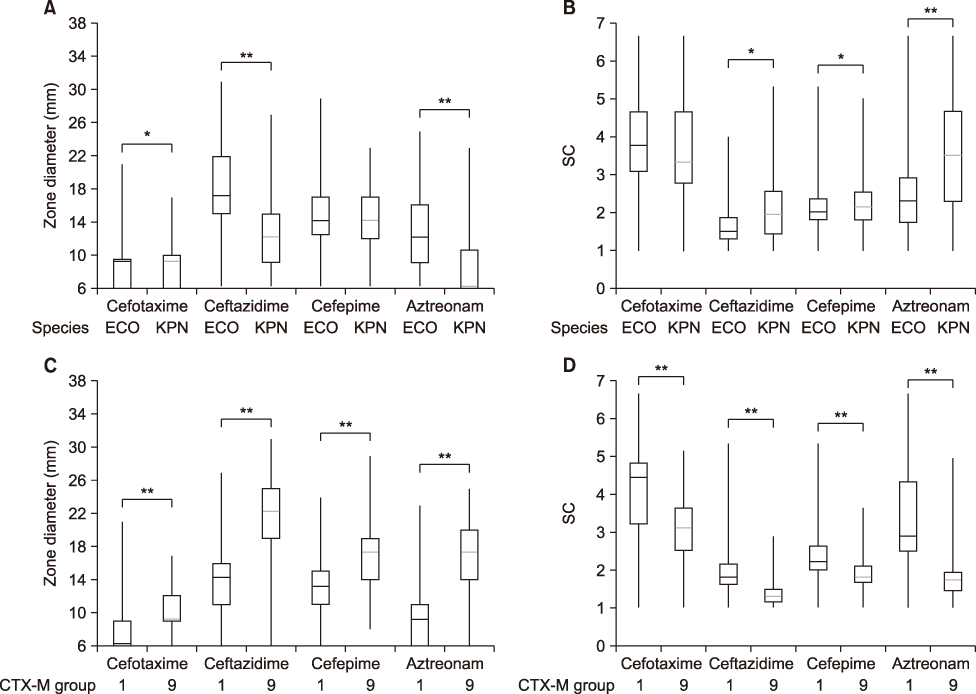
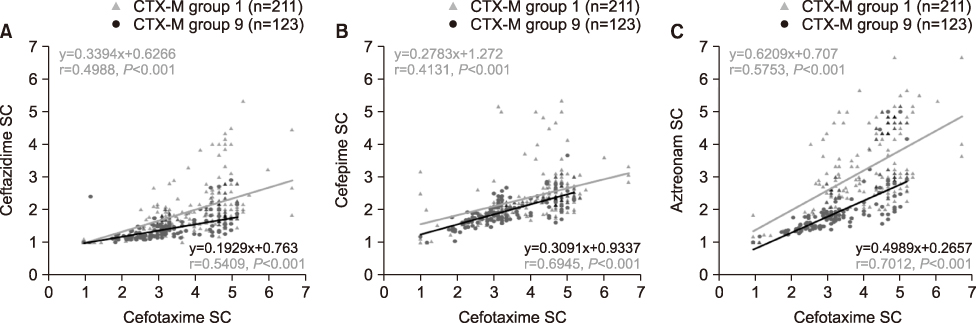
Of total microbes, 31.6% (303/959) E. coli and 24.0% (91/379) K. pneumoniae were resistant to cefotaxime and 28.1% (269/959) E. coli and 20.1% (76/379) K. pneumoniae were CTX-M-type ESBL producers. Among the detected CTX-M ESBLs, 58.0% (156/269) in E. coli and 86.8% (66/76) in K. pneumoniae belonged to group 1, 46.8% (126/269) in E. coli and 14.5% (11/76) in K. pneumoniae were group 9. Ten E. coli and one K. pneumoniae isolates co-produced both groups of CTX-M ESBL. The group 1 CTX-M producers had a higher level of resistance to cefotaxime, ceftazidime, cefepime, and aztreonam and exhibited stronger synergistic activities when combined with CA compared to group 9.
CONCLUSION
ESBL phenotypes differ by CTX-M ESBL group and phenotype testing with drugs including 4th generation cephalosporins and monobactams is critical for screening CTX-M-producers with better sensitivity.
MeSH Terms
Figure
Reference
-
1. Pitout JD. Extended-spectrum beta-lactamaseproducing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008; 8:159–166.2. Paterson DL. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005; 18:657–686.3. D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013; 303:305–317.4. Matsumoto Y, Ikeda F, Kamimura T, Yokota Y, Mine Y. Novel plasmid-mediated beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob Agents Chemother. 1988; 32:1243–1246.5. Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990; 18:294–298.6. Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004; 48:1–14.7. Delmas J, Leyssene D, Dubois D, Birck C, Vazeille E, Robin F, et al. Structural insights into substrate recognition and product expulsion in CTX-M enzymes. J Mol Biol. 2010; 400:108–120.
Article8. Livermore DM. Determinants of the activity of beta-lactamase inhibitor combinations. J Antimicrob Chemother. 1993; 31:Suppl A. 9–21.9. Livermore DM, Hope R, Mushtaq S, Warner M. Orthodox and unorthodox clavulanate combinations against extended-spectrum beta-lactamase producers. Clin Microbiol Infect. 2008; 14:Suppl 1. 189–193.10. Livermore DM, Akova M, Wu PJ, Yang YJ. Clavulanate and beta-lactamase induction. J Antimicrob Chemother. 1989; 24:Suppl B. 23–33.11. CLSI. CLSI document M100-S25. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute;2015.12. European Committee on Antimicrobial Susceptibility Testing. EUCAST Guideline for the Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Version 2.0. Vasel: EUCAST;2017.13. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011; 70:119–123.
Article14. Ryoo NH, Kim EC, Hong SG, Park YJ, Lee K, Bae IK, et al. Dissemination of SHV-12 and CTX-M-type extended-spectrum beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 2005; 56:698–702.15. Pérez-Pérez FJ. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002; 40:2153–2162.16. Campbell MJ. Statistics at Square Two. London: BMJ Books;2006.17. Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017; 72:2145–2155.
Article18. Kim D, Ahn JY, Lee CH, Jang SJ, Lee H, Yong D, et al. Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in Gram-negative bacilli and the emergence of carbapenem non-susceptibility in Klebsiella pneumoniae: analysis of Korean Antimicrobial Resistance Monitoring System (KARMS) data from 2013 to 2015. Ann Lab Med. 2017; 37:231–239.19. Park SH, Byun JH, Choi SM, Lee DG, Kim SH, Kwon JC, et al. Molecular epidemiology of extended-spectrum β-lactamaseproducing Escherichia coli in the community and hospital in Korea: emergence of ST131 producing CTX-M-15. BMC Infect Dis. 2012; 12:149.
Article20. Stedt J, Bonnedahl J, Hernandez J, Waldenström J, McMahon BJ, Tolf C, et al. Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand. 2015; 57:74.
Article21. Timofte D, Maciuca IE, Williams NJ, Wattret A, Schmidt V. Veterinary hospital dissemination of CTX-M-15 extended-spectrum beta-lactamase-producing Escherichia coli ST410 in the United Kingdom. Microb Drug Resist. 2016; 22:609–615.22. Ojer-Usoz E, González D, Vitas AI. Clonal diversity of ESBL-producing Escherichia coli isolated from environmental, human and food samples. Int J Environ Res Public Health. 2017; 14:E676.23. Shin SW, Jung M, Won HG, Belaynehe KM, Yoon IJ, Yoo HS. Characteristics of transmissible CTX-M- and CMY-Type β-lactamase-producing Escherichia coli isolates collected from pig and chicken farms in South Korea. J Microbiol Biotechnol. 2017; 27:1716–1723.24. Kim HS, Chon JW, Kim YJ, Kim DH, Kim MS, Seo KH. Prevalence and characterization of extended-spectrum-β-lactamaseproducing Escherichia coli and Klebsiella pneumoniae in readyto-eat vegetables. Int J Food Microbiol. 2015; 207:83–86.25. Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966; 45:493–496.
Article26. Maurer FP, Courvalin P, Böttger EC, Hombach M. Integrating forecast probabilities in antibiograms: a way to guide antimicrobial prescriptions more reliably? J Clin Microbiol. 2014; 52:3674–3684.
Article27. Bonnet R, Recule C, Baraduc R, Chanal C, Sirot D, De Champs C, et al. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother. 2003; 52:29–35.
Article28. Poirel L, Gniadkowski M, Nordmann P. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum beta-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J Antimicrob Chemother. 2002; 50:1031–1034.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Significance of Extended-spectrum β-lactamase-producing Bacteria in First Pediatric Febrile Urinary Tract Infections and Differences between Age Groups
- Diversity of Genetic Environment of bla(CTX-M) Genes and Antimicrobial Susceptibility in Extended-spectrum β-lactamase producing Escherichia coli Isolated in Korea
- Characterization of Extended-Spectrum β-Lactamase Genes of Shigella flexneri Isolates With Fosfomycin Resistance From Patients in China
- The Emerging Strong Among Extended-Spectrum beta-Lactamases: CTX-M Enzymes
- Infection of Extended-Spectrum β-Lactamase Producing Shigella flexneri in Children Attending a Childcare Center in Korea