J Bacteriol Virol.
2019 Sep;49(3):95-114. 10.4167/jbv.2019.49.3.95.
Diversity of Genetic Environment of bla(CTX-M) Genes and Antimicrobial Susceptibility in Extended-spectrum β-lactamase producing Escherichia coli Isolated in Korea
- Affiliations
-
- 1Department of Microbiology, Keimyung University School of Medicine, 1035 Dalgubeol-daero, Dalseo-Gu, Daegu, 42601, Korea. minho@dsmc.or.kr
- KMID: 2468030
- DOI: http://doi.org/10.4167/jbv.2019.49.3.95
Abstract
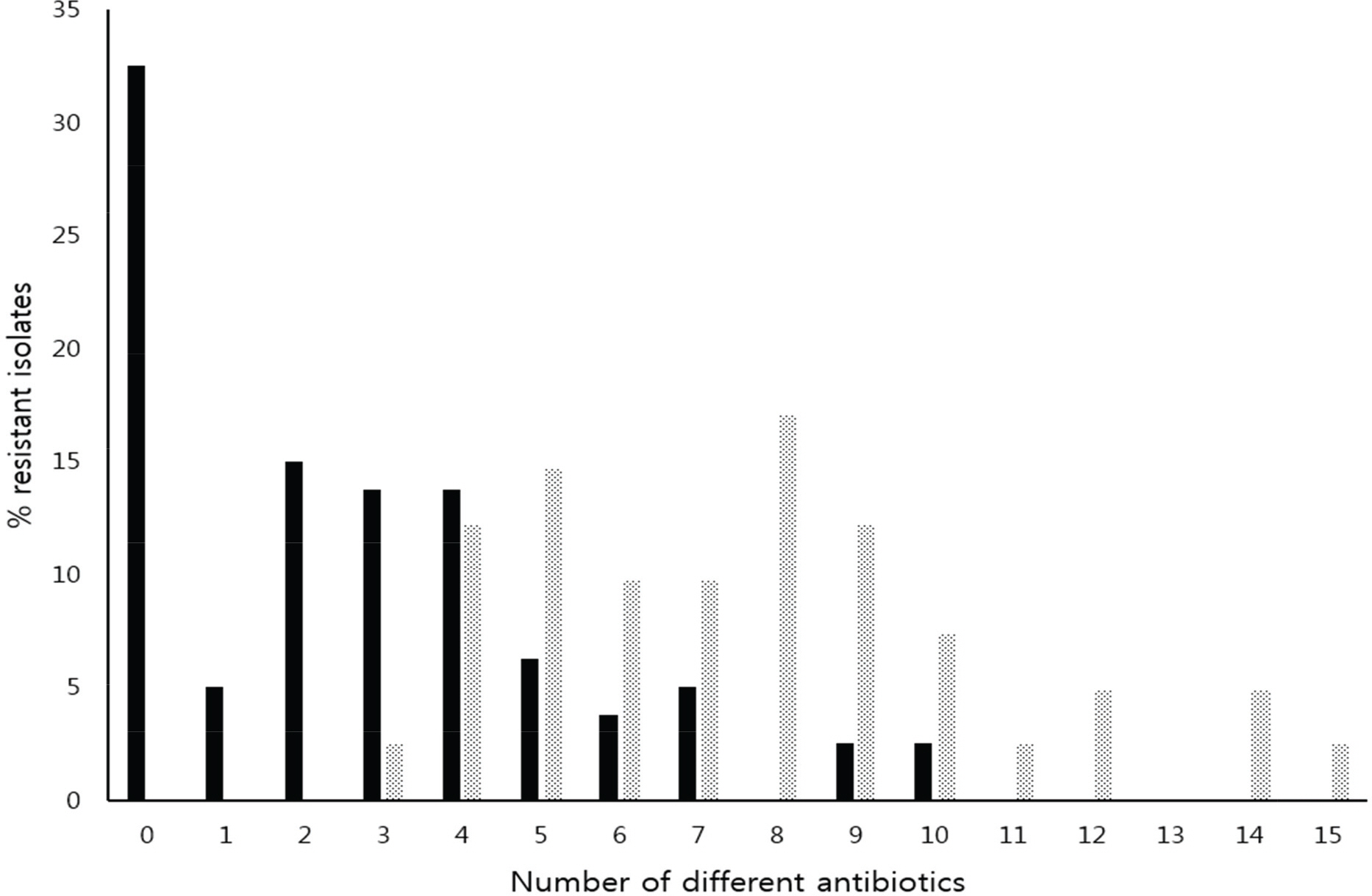
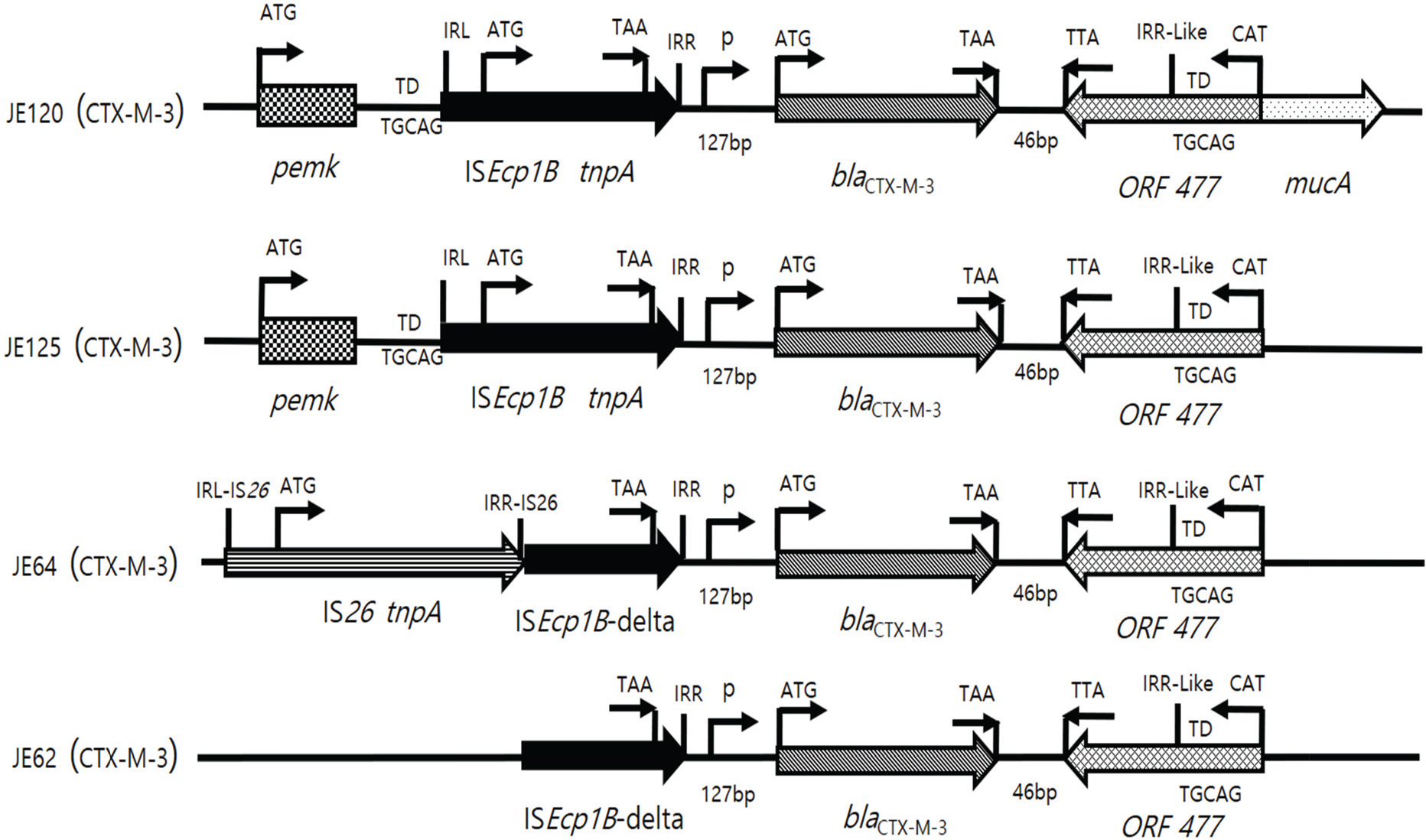
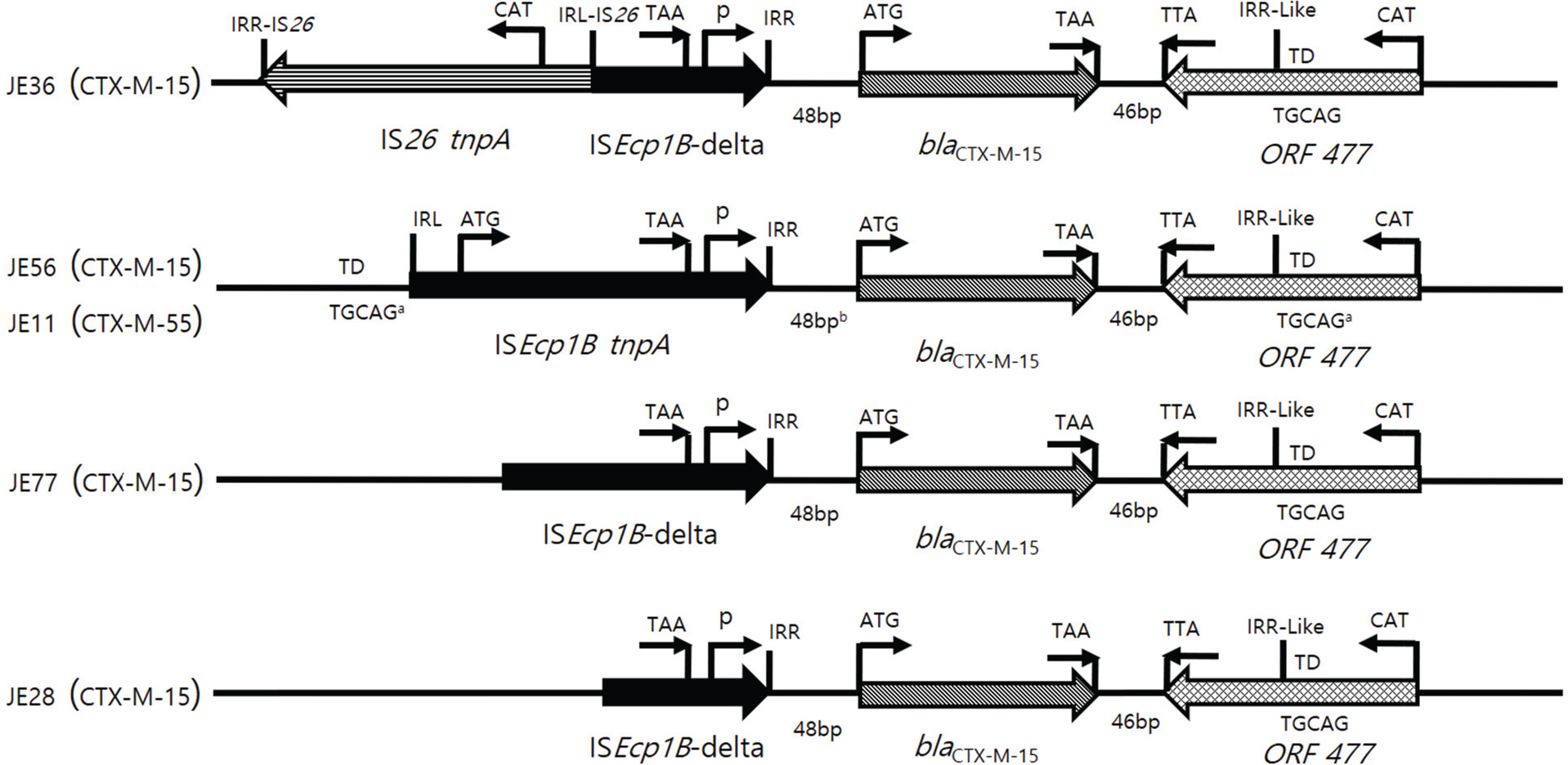
- Increasing resistance due to the production of extended-spectrum β-lactamase (ESBL) in Escherichia coli is a major problem to public health and CTX-M enzymes have become the most prevalent ESBL worldwide. In this study, resistance profiles of E. coli isolated in Korea and the genetic environments of bla(CTX-M) genes were analyzed by PCR and direct sequencing to clarify the mechanisms of spread of CTX-M. Resistance rates of CTX-M-producing E. coli, including β-lactams, fluoroquinolones and aminoglycosides, were significantly higher than that of CTX-M-non-producers (p<0.01). Of 41 tested, 39 (95.1%) isolates of CTX-M-producing E. coli showed resistance transfer by conjugation. All the transconjugants harboured large plasmids of 118~172 megadalton. Insertion sequence ISEcp1B was detected in the upstream of the bla(CTX-M) in 38 (92.7%) isolates with bla(CTX-M). ISEcp1B was disrupted by IS26 in 16 (39.0%) isolates with bla(CTX-M). ISEcp1B carried −35 and −10 promoter components between right inverted repeat (IRR) and the start codon of bla(CTX-M). orf477 or IS903D was observed in the downstream of the bla(CTX-M) in all the isolates with bla(CTX-M-3/15/55) or with bla(CTX-M-14/27), respectively. Sequence similar to IRR of ISEcp1B was located downstream of orf477. Target duplication sequences were detected both upstream of IRL and downstream of IRR. These results showed the involvement of ISEcp1B in the mobilization of the resistance genes. In conclusion, the surrounding DNAs of bla(CTX-M) genes were very diverse, and the spread and the expression of CTX-M may be deeply related with ISEcp1B. These informations will provide important knowledge to control the increase in CTX-M-ESBLs.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Analysis of Integrons and Antimicrobial Resistances of Multidrug Resistant Escherichia coli Isolated in Korea
Yun-Yi Yang, Min-Ho Suh
J Bacteriol Virol. 2019;49(4):176-190. doi: 10.4167/jbv.2019.49.4.176.
Reference
-
1). Rossolini GM, D’ Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum β-lactamases. Clin Microbiol Infect. 2008; 14:33–41.
Article2). Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010; 13:558–64.3). Pitout JD, Laupland KB. Extendedspectrum β- lactamase- producing Enterobacteriaceae: an emerging public- health concern. Lancet Infect Dis. 2008; 8:159–66.4). Pottinger P, Reller LB, Ryan KJ. Enterobacteriaceae. Ryan KJ, Ray CG, editors. Sherris Medical Microbiology. 6th ed.New York: McGraw-Hill Education, Inc.;2014. p. 579–608.5). Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011; 66:1–14.6). Naseer U, Sundsfjord A. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist. 2011; 17:83–97.7). Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009; 48:1–12.
Article8). Kang CI, Wi YM, Lee MY, Ko KS, Chung DR, Peck KR, et al. Epidemiology and risk factors of community onset infections caused by extended-spectrum β-lactamase-producing Escherichia coli strains. J Clin Microbiol. 2012; 50:312–7.9). El Salabi A, Walsh TR, Chouchani C. Extended spectrum betalactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gramnegative bacteria. Crit Rev Microbiol. 2013; 39:113–22.10). Skippington E, Ragan MA. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011; 35:707–35.
Article11). Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013; 303:298–304.
Article12). Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. 2014; 38:865–91.
Article13). Hickman AB, Dyda F. Mechanisms of DNA transposition. Craig NL, editor. Mobile DNA III. 3rd ed.Washington, DC: ASM Press;2015. p. 531–53.
Article14). Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. Everyman's guide to bacterial insertion sequences. Craig NL, editor. Mobile DNA III. 3rd ed.Washington, DC: ASM Press;2015. p. 555–90.
Article15). Escudero JA, Loot C, Nivina A, Mazel D. The Integron: Adaptation on demand. Craig NL, editor. Mobile DNA III. 3rd ed.Washington, DC: ASM Press;2015. p. 139–61.16). Lartigue MF, Poirel L, Nordmann P. Diversity of genetic environment of bla CTX- M genes. FEMS Microbiol Lett. 2004; 234:201–7.17). Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various bla CTX- M genes. J Antimicrob Chemother. 2006; 57:14–23.18). Zhao WH, Hu ZQ. Epidemiology and genetics of CTX-M extended- spectrum β- lactamases in Gram-negative bacteria. Crit Rev Microbiol. 2013; 39:79–101.19). Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, et al. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother. 2015; 70:1639–49.20). Strockbine NA, Bopp CA, Fields PI, Kaper JB, Nataro JP. Escherichia, Shigella, and Salnomella. Jorgensen JH, Pfaller MA, editors. Manual of Clinical Microbiology. 11th ed.Washington, DC: ASM Press;2015. p. 685–713.21). Jorgensen JH, Turnidge JD. Susceptibility test methods: Dilution and disk diffusion methods. Jorgensen JH, Pfaller MA, editors. Manual of Clinical Microbiology. 11th ed.Washington, DC: ASM Press;2015. p. 1253–73.
Article22). Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 24th ed.Wayne: CLSI Press;2014.23). Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug- resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18:268–81.24). Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981; 145:1365–73.
Article25). Levy-Hara G, Amá bile-Cuevas CF, Gould I, Hutchinson J, Abbo L, Saxynger L, et al. “Ten commandments” for the appropriate use of antibiotics by the practicing physician in an outpatient setting. Front Microbiol. 2011; 2:230.
Article26). Matsumura Y, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob Agents Chemother. 2013; 57:4736–42.27). Shin J, Kim DH, Ko KS. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J Infect. 2011; 63:39–47.28). Bonnet R, Recule C, Baraduc R, Chanal C, Sirot D, De Champs C, et al. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother. 2003; 52:29–35.
Article29). Cantó n R, Morosini M. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev. 2011; 35:977–91.
Article30). Amá bile-Cuevas CF. Antibiotic resistance: From Darwin to Lederberg to Keynes. Microb Drug Resist. 2013; 19:73–87.31). Kuo HY, Chang KC, Kuo JW, Yueh HW, Liou ML. Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 2012; 39:33–8.32). Hegstad K, Samuelsen O, Hegstad J, Sundsfjord A. Molecular methods for detection of antibacterial resistance genes: rationale and applications. Amsterdam D, editor. Antibiotics in Laboratory Medicine. 6th ed.Philadelphia: Wolters Kluwer Health;2015. p. 408–49.33). Adler A, Gniadkowski M, Baraniak A, Izdebski R Fiett J, Hryniewicz W, et al. Transmission dynamics of ESBL-producing Escherichia coli clones in rehabilitation wards at a tertiary care centre. Clin Microbiol Infect. 2012; 18:497–505.34). Ruiz del Castillo B, Vinué L, Romá n EJ, Guerra B, Carattoli A, Torres C, et al. Molecular characterization of multiresistant Escherichia coli producing or not extended- spectrum β- lactamases. BMC Microbiol. 2013; 13:84.35). Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. Characterization of IncF plasmids carrying the bla CTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother. 2011; 66:1263–8.36). Matsumura Y, Yamamoto M, Nagao M, Hotta G, Matsushima A, Ito Y, et al. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-β-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother. 2012; 67:2612–20.37). Mnif B, Harhour H, Jdidi J, Mahjoubi F, Genel N, Arlet G, et al. Molecular epidemiology of extended- spectrum β- lactamase- producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol. 2013; 13:147.38). Poirel L, Lartigue MF, Decousser JW, Nordmann P. ISEcp1B- mediated transposition of bla CTX- M in Escherichia coli. Antimicrob Agents Chemother. 2005; 49:447–50.39). Readman JB, Dickson G, Coldham NG. Translational inhibition of CTX-M extended- spectrum β- lactamase in clinical strains of Escherichia coli by synthetic antisense oligonucleotides partially restores sensitivity to cefotaxime. Front Microbiol. 2016; 7:373.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Comparisons of CTX-M-Producing Escherichia coli Isolates from Humans and Animals in South Korea
- Molecular characteristics of ESBLproducing Escherichia coli isolated from chickens with colibacillosis
- In Vitro Susceptibility of piperacillin/tazobactam Against extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae
- Prevalence of Fecal Carriage of CTX-M-15 Beta-Lactamase-Producing Escherichia coli in Healthy Children from a Rural Andean Village in Venezuela
- Prevalence of Ambler Class A Extended-Spectrum beta-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae Isolates in Korea