J Breast Cancer.
2018 Sep;21(3):330-333. 10.4048/jbc.2018.21.e33.
The Novel Pathogenic Mutation c.849dupT in BRCA2 Contributes to the Nonsense-Mediated mRNA Decay of BRCA2 in Familial Breast Cancer
- Affiliations
-
- 1Wuhan Red Cross Hospital, Wuhan, China. 1451727512@qq.com
- 2Shanghai Key Laboratory of Birth Defect, Children's Hospital of Fudan University, Shanghai, China.
- 3Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, Fudan University, Shanghai, China.
- 4Department of Biochemistry and Molecular Biology, Collaborative Innovation Center of Genetics and Development, Institutes of Biomedical Sciences, School of Basic Medical Sciences, Fudan University, Shanghai, China.
- KMID: 2421374
- DOI: http://doi.org/10.4048/jbc.2018.21.e33
Abstract
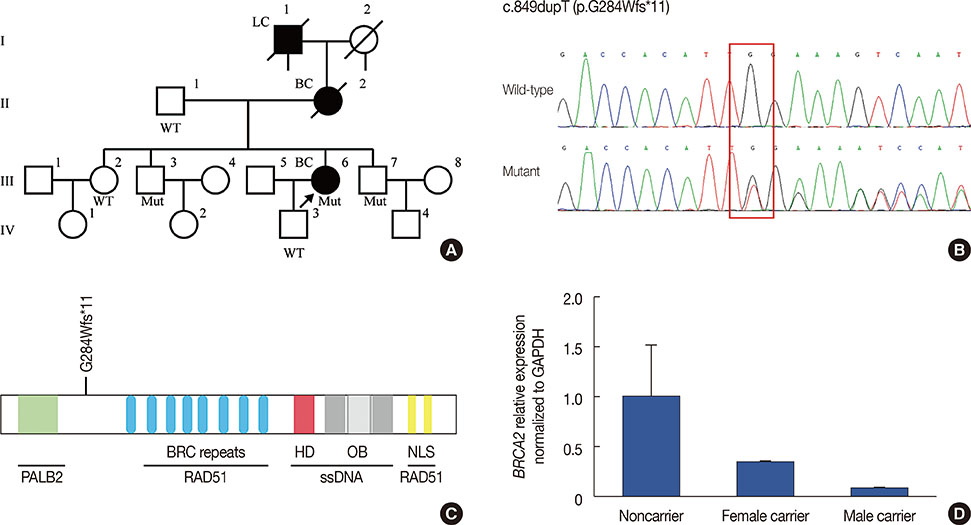
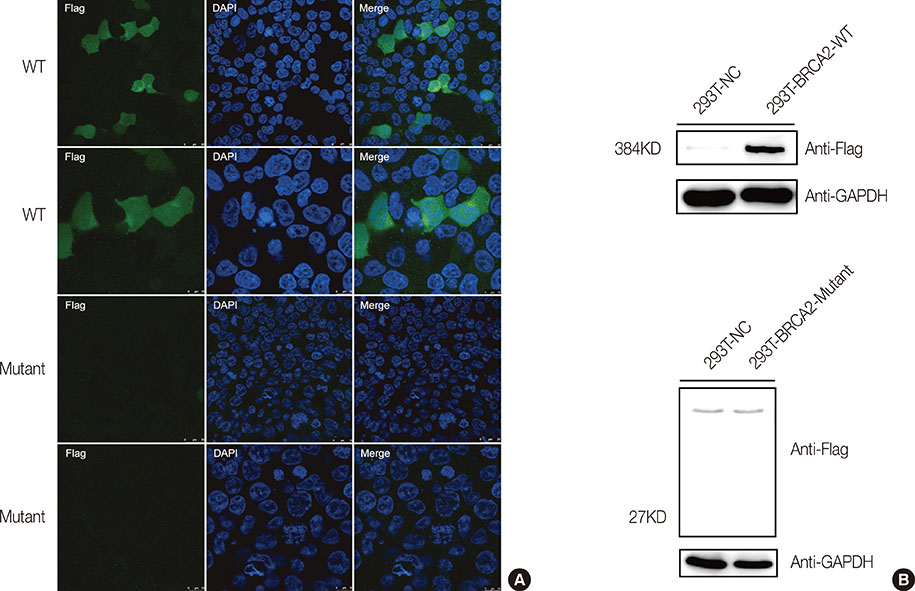
- In this study, we used next-generation sequencing methods to screen 300 individuals for BRCA1 and BRCA2. A novel mutation (c.849dupT) in BRCA2 was identified in a female patient and her unaffected brothers. This mutation leads to the truncation of BRCA2 functional domains. Moreover, BRCA2 mRNA expression levels in mutation carriers are significantly reduced compared to noncarriers. Immunofluorescence and western blot assays showed that this mutation resulted in reduced BRCA2 protein expression. Thus, we identified a novel mutation that damaged the function and expression of BRCA2 in a family with breast cancer history. The pedigree analysis suggested that this mutation is strongly associated with familial breast cancer. Genetic counsellors suggest that mutation carriers in this family undergo routine screening for breast cancer, as well as other malignancies, such as prostate and ovarian cancer. The effects of this BRCA2 mutation on drug resistance should be taken into consideration during treatment.
Keyword
MeSH Terms
Figure
Reference
-
1. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136:E359–E386.
Article2. Ly D, Forman D, Ferlay J, Brinton LA, Cook MB. An international comparison of male and female breast cancer incidence rates. Int J Cancer. 2013; 132:1918–1926.
Article3. Patel UA, Perry M, Crane-Robinson C. Screening for germline mutations of the p53 gene in familial breast cancer patients. Eur J Clin Invest. 1995; 25:132–137.
Article4. Apostolou P, Papasotiriou I. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer (Dove Med Press). 2017; 9:331–335.
Article5. Cao WM, Gao Y, Yang HJ, Xie SN, Ding XW, Pan ZW, et al. Novel germline mutations and unclassified variants of BRCA1 and BRCA2 genes in Chinese women with familial breast/ovarian cancer. BMC Cancer. 2016; 16:64.
Article6. Ma J, Yang J, Jian W, Wang X, Xiao D, Xia W, et al. A novel loss-of-function heterozygous BRCA2 c.8946_8947delAG mutation found in a Chinese woman with family history of breast cancer. J Cancer Res Clin Oncol. 2017; 143:631–637.
Article7. Hofstatter EW, Domchek SM, Miron A, Garber J, Wang M, Componeschi K, et al. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer. 2011; 10:225–231.
Article8. Ouhtit A, Gupta I, Shaikh Z. BRIP1, a potential candidate gene in development of non-BRCA1/2 breast cancer. Front Biosci (Elite Ed). 2016; 8:289–298.
Article9. Antoniou AC, Easton DF. Models of genetic susceptibility to breast cancer. Oncogene. 2006; 25:5898–5905.
Article10. Skol AD, Sasaki MM, Onel K. The genetics of breast cancer risk in the post-genome era: thoughts on study design to move past BRCA and towards clinical relevance. Breast Cancer Res. 2016; 18:99.
Article11. Bayraktar S, Arun B. BRCA mutation genetic testing implications in the United States. Breast. 2017; 31:224–232.
Article12. Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002; 297:1837–1848.
Article13. Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat Genet. 1996; 13:22–23.
Article14. Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014; 15:585–598.
Article15. Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003; 17:3017–3022.
Article16. Min J, Choi ES, Hwang K, Kim J, Sampath S, Venkitaraman AR, et al. The breast cancer susceptibility gene BRCA2 is required for the maintenance of telomere homeostasis. J Biol Chem. 2012; 287:5091–5101.
Article17. Nientiedt C, Heller M, Endris V, Volckmar AL, Zschäbitz S, Tapia-Laliena MA, et al. Mutations in BRCA2 and taxane resistance in prostate cancer. Sci Rep. 2017; 7:4574.
Article18. Dhillon KK, Swisher EM, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011; 102:663–669.
Article19. Basham VM, Lipscombe JM, Ward JM, Gayther SA, Ponder BA, Easton DF, et al. BRCA1 and BRCA2 mutations in a population-based study of male breast cancer. Breast Cancer Res. 2002; 4:R2.
Article20. Couch FJ, Farid LM, DeShano ML, Tavtigian SV, Calzone K, Campeau L, et al. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet. 1996; 13:123–125.
Article21. Castro E, Goh C, Olmos D, Saunders E, Leongamornlert D, Tymrakiewicz M, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013; 31:1748–1757.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- Novel Germline Mutations of BRCA1 and BRCA2 in Korean Familial Breast Cancer Patients
- Bilateral Triple-Negative Invasive Breast Cancer with a BRCA2 Mutation, and Glioblastoma: A Case Report and Literature Review
- BRCA1/BRCA2 Pathogenic Variant Breast Cancer: Treatment and Prevention Strategies
- Endometrial cancer occurence five years after breast cancer in BRCA2 mutation patient